Gene
KWMTBOMO06244 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001806
Annotation
cleavage_and_polyadenylation_specific_factor_5_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.136 Mitochondrial Reliability : 1.066 Nuclear Reliability : 1.205
Sequence
CDS
ATGAATTTAACTCTCAATAGGTCTATTAATTTGTATCCGCTTACGAATTACACGTTCGGAACTAAGGAGCCGCTGTTCGAGAAAGACGCGTCGGTCCCGGCGAGGTTCCAGAGGATGCGAGAGGAGTTTTGTAAAATCGGAATGAGAAGGTCAGTCGAAGGTGTGTTATTAGTACATGAACATGGTCTCCCTCATGTGCTGTTACTGCAACTCGGCACTGCGTTTTTCAAGTTGCCCGGTGGAGAATTGAATCCTGGAGAGGATGAAATTGATGGTTTGAAGAGGCTTCTCACAGAGACTTTAGGCAGGCAAGACGGTGTCAAACAGGAATGGCTCATTGAAGACACAATAGGAAACTGGTGGCGACCGAATTTCGAACCACCGCAATATCCATACATTCCGCCTCACATCACGAAACCAAAGGAGCATAAGCGCCTATTCCTGGTTCAACTGCAAGACAGAGCTCTGTTCGCTGTACCGAAGAATTACAAGCTGGTTGCGGCACCATTGTTTGAATTGTACGATAATGCTCAGGGATACGGACCTATAATATCGTCGTTGTCTCAGAGTTTATGTCGTTTTAATTTCATTTATATGTGA
Protein
MNLTLNRSINLYPLTNYTFGTKEPLFEKDASVPARFQRMREEFCKIGMRRSVEGVLLVHEHGLPHVLLLQLGTAFFKLPGGELNPGEDEIDGLKRLLTETLGRQDGVKQEWLIEDTIGNWWRPNFEPPQYPYIPPHITKPKEHKRLFLVQLQDRALFAVPKNYKLVAAPLFELYDNAQGYGPIISSLSQSLCRFNFIYM
Summary
Uniprot
Q1HQ96
S4P3R5
A0A1E1WIP5
A0A2W1BQ18
A0A2A4J8P7
A0A2H1WRK1
+ More
A0A067QW03 A0A1B6EAM4 E2C6H0 A0A0L7QWX6 A0A2A3EQM6 V9IF30 A0A088A0J5 A0A154P8R7 A0A195CYZ3 A0A195FWH7 A0A151WVX6 A0A026WUE0 A0A151IXD2 F4WUX3 A0A195AUB3 A0A158P0R1 A0A0R3P7I2 A0A1I8PR01 A0A1I8ML55 A0A2C9GRS2 Q7QJ32 A0A0Q9WZL1 A0A1B6MAG1 A0A0J9RSI2 Q0E8G6 A0A0R1DUQ0 A0A0Q5U3M9 A0A182JJZ8 A0A1W4VM99 A0A182MBC9 A0A1B6EQV4 A0A1B6JV95 A0A0P9C4Y2 A0A0K8TRK9 A0A1L8DGK5 A0A1B0CSL1 A0A182G2Q1 Q17DC9 A0A3B0JP98 A0A336M9B6 A0A2M4CT68 A0A2M4AXE1 A0A0Q9WIP5 A0A0Q9XMR2 A0A1Q3G177 A0A069DXT7 A0A1L8DGU6 A0A0P4VII3 A0A0K8W3P3 A0A0A1WSL8 A0A034WNR1 A0A0M5J0W6 U5EVX9 A0A0T6B375 A0A0C9S034 A0A2P8Z8W4 A0A1Y1K6G0 A0A182XLL2 E0VW43 A0A232ENQ1 D7EI10 A0A023F8J5 A0A182YMM7 R4FKV7 A0A293LYT4 A0A2J7QTZ6 A0A0V0GAF0 A0A069DPX3 A0A2R5LGY6 A0A131XD89 A0A131YR84 L7ME45 A0A0A9VUK6 A0A182NKR8 A0A023GGI1 A0A1E1XBK4 K7ISQ4 A0A0P5VTV5 E2AFE5 G3MM11 A0A0N8DXN1 A0A0P4YBY5 A0A0P6J4F1 A0A0P5S419 E9HF00 A0A087T658 A0A1A9WTL7 A0A0P6EY85 A0A0P5MVV3 A0A1A9Y0S4 D3TNW4 A0A0P5PLN6 A0A0P5V6E2 A0A0P5V2A3
A0A067QW03 A0A1B6EAM4 E2C6H0 A0A0L7QWX6 A0A2A3EQM6 V9IF30 A0A088A0J5 A0A154P8R7 A0A195CYZ3 A0A195FWH7 A0A151WVX6 A0A026WUE0 A0A151IXD2 F4WUX3 A0A195AUB3 A0A158P0R1 A0A0R3P7I2 A0A1I8PR01 A0A1I8ML55 A0A2C9GRS2 Q7QJ32 A0A0Q9WZL1 A0A1B6MAG1 A0A0J9RSI2 Q0E8G6 A0A0R1DUQ0 A0A0Q5U3M9 A0A182JJZ8 A0A1W4VM99 A0A182MBC9 A0A1B6EQV4 A0A1B6JV95 A0A0P9C4Y2 A0A0K8TRK9 A0A1L8DGK5 A0A1B0CSL1 A0A182G2Q1 Q17DC9 A0A3B0JP98 A0A336M9B6 A0A2M4CT68 A0A2M4AXE1 A0A0Q9WIP5 A0A0Q9XMR2 A0A1Q3G177 A0A069DXT7 A0A1L8DGU6 A0A0P4VII3 A0A0K8W3P3 A0A0A1WSL8 A0A034WNR1 A0A0M5J0W6 U5EVX9 A0A0T6B375 A0A0C9S034 A0A2P8Z8W4 A0A1Y1K6G0 A0A182XLL2 E0VW43 A0A232ENQ1 D7EI10 A0A023F8J5 A0A182YMM7 R4FKV7 A0A293LYT4 A0A2J7QTZ6 A0A0V0GAF0 A0A069DPX3 A0A2R5LGY6 A0A131XD89 A0A131YR84 L7ME45 A0A0A9VUK6 A0A182NKR8 A0A023GGI1 A0A1E1XBK4 K7ISQ4 A0A0P5VTV5 E2AFE5 G3MM11 A0A0N8DXN1 A0A0P4YBY5 A0A0P6J4F1 A0A0P5S419 E9HF00 A0A087T658 A0A1A9WTL7 A0A0P6EY85 A0A0P5MVV3 A0A1A9Y0S4 D3TNW4 A0A0P5PLN6 A0A0P5V6E2 A0A0P5V2A3
Pubmed
19121390
23622113
28756777
24845553
20798317
24508170
+ More
30249741 21719571 21347285 15632085 17994087 25315136 12364791 14747013 17210077 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 26369729 26483478 17510324 26334808 27129103 25830018 25348373 29403074 28004739 20566863 28648823 18362917 19820115 25474469 25244985 28049606 26830274 25576852 25401762 26823975 28503490 20075255 22216098 21292972 20353571
30249741 21719571 21347285 15632085 17994087 25315136 12364791 14747013 17210077 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 26369729 26483478 17510324 26334808 27129103 25830018 25348373 29403074 28004739 20566863 28648823 18362917 19820115 25474469 25244985 28049606 26830274 25576852 25401762 26823975 28503490 20075255 22216098 21292972 20353571
EMBL
BABH01021254
DQ443156
ABF51245.1
GAIX01006329
JAA86231.1
GDQN01004288
+ More
JAT86766.1 KZ150084 PZC73793.1 NWSH01002344 PCG68487.1 ODYU01010528 SOQ55700.1 KK852893 KDR14219.1 GEDC01002335 JAS34963.1 GL453156 EFN76452.1 KQ414705 KOC63128.1 KZ288201 PBC33572.1 JR040285 AEY59066.1 KQ434846 KZC08262.1 KQ977141 KYN05354.1 KQ981200 KYN45020.1 KQ982691 KYQ52044.1 KK107102 QOIP01000005 EZA59622.1 RLU22870.1 KQ980819 KYN12684.1 GL888378 EGI61987.1 KQ976738 KYM75647.1 ADTU01005767 ADTU01005768 CH379067 KRT07411.1 APCN01000179 AAAB01008807 EAA04203.3 CH963847 KRF97624.1 GEBQ01007071 JAT32906.1 CM002912 KMY98632.1 AE014296 ABI31246.1 CM000159 KRK00876.1 CH954178 KQS43695.1 AXCM01002371 GECZ01029428 GECZ01018463 JAS40341.1 JAS51306.1 GECU01030530 GECU01013832 GECU01004575 JAS77176.1 JAS93874.1 JAT03132.1 CH902618 KPU78791.1 GDAI01000827 JAI16776.1 GFDF01008498 JAV05586.1 AJWK01026189 AJWK01026190 AJWK01026191 JXUM01039933 KQ561224 KXJ79300.1 CH477297 CH477216 EAT44361.1 EAT47515.1 OUUW01000002 SPP77320.1 UFQS01000716 UFQS01000742 UFQT01000716 UFQT01000742 SSX06379.1 SSX26866.1 GGFL01004291 MBW68469.1 GGFK01012133 MBW45454.1 CH940647 KRF84472.1 CH933809 KRG06206.1 GFDL01001505 JAV33540.1 GBGD01002715 JAC86174.1 GFDF01008499 JAV05585.1 GDKW01001974 JAI54621.1 GDHF01006632 JAI45682.1 GBXI01012495 JAD01797.1 GAKP01002678 GAKP01002677 JAC56274.1 CP012525 ALC43636.1 GANO01001671 JAB58200.1 LJIG01016003 KRT81907.1 GBYB01013759 GBYB01013761 JAG83526.1 JAG83528.1 PYGN01000143 PSN52932.1 GEZM01093979 JAV56038.1 DS235817 EEB17599.1 NNAY01003100 OXU19980.1 KQ972204 EFA12938.2 GBBI01001011 JAC17701.1 ACPB03017369 ACPB03017370 GAHY01001683 JAA75827.1 GFWV01008282 MAA33011.1 NEVH01011192 PNF32060.1 GECL01001219 JAP04905.1 GBGD01002761 JAC86128.1 GGLE01004667 MBY08793.1 GEFH01004513 JAP64068.1 GEDV01007459 JAP81098.1 GACK01002962 JAA62072.1 GBHO01044215 GBRD01005297 GDHC01014014 JAF99388.1 JAG60524.1 JAQ04615.1 GBBM01002366 JAC33052.1 GFAC01002564 JAT96624.1 AAZX01007951 GDIP01095718 JAM07997.1 GL439108 EFN67838.1 JO842912 AEO34529.1 GDIQ01081820 JAN12917.1 GDIP01233302 JAI90099.1 GDIQ01013402 JAN81335.1 GDIQ01152793 GDIQ01092666 GDIP01074909 GDIQ01031437 JAL59060.1 JAM28806.1 GL732632 EFX69694.1 KK113612 KFM60597.1 GDIQ01056879 JAN37858.1 GDIQ01152792 JAK98933.1 CCAG010000251 EZ423116 ADD19392.1 GDIQ01137151 JAL14575.1 GDIP01119661 JAL84053.1 GDIP01121654 JAL82060.1
JAT86766.1 KZ150084 PZC73793.1 NWSH01002344 PCG68487.1 ODYU01010528 SOQ55700.1 KK852893 KDR14219.1 GEDC01002335 JAS34963.1 GL453156 EFN76452.1 KQ414705 KOC63128.1 KZ288201 PBC33572.1 JR040285 AEY59066.1 KQ434846 KZC08262.1 KQ977141 KYN05354.1 KQ981200 KYN45020.1 KQ982691 KYQ52044.1 KK107102 QOIP01000005 EZA59622.1 RLU22870.1 KQ980819 KYN12684.1 GL888378 EGI61987.1 KQ976738 KYM75647.1 ADTU01005767 ADTU01005768 CH379067 KRT07411.1 APCN01000179 AAAB01008807 EAA04203.3 CH963847 KRF97624.1 GEBQ01007071 JAT32906.1 CM002912 KMY98632.1 AE014296 ABI31246.1 CM000159 KRK00876.1 CH954178 KQS43695.1 AXCM01002371 GECZ01029428 GECZ01018463 JAS40341.1 JAS51306.1 GECU01030530 GECU01013832 GECU01004575 JAS77176.1 JAS93874.1 JAT03132.1 CH902618 KPU78791.1 GDAI01000827 JAI16776.1 GFDF01008498 JAV05586.1 AJWK01026189 AJWK01026190 AJWK01026191 JXUM01039933 KQ561224 KXJ79300.1 CH477297 CH477216 EAT44361.1 EAT47515.1 OUUW01000002 SPP77320.1 UFQS01000716 UFQS01000742 UFQT01000716 UFQT01000742 SSX06379.1 SSX26866.1 GGFL01004291 MBW68469.1 GGFK01012133 MBW45454.1 CH940647 KRF84472.1 CH933809 KRG06206.1 GFDL01001505 JAV33540.1 GBGD01002715 JAC86174.1 GFDF01008499 JAV05585.1 GDKW01001974 JAI54621.1 GDHF01006632 JAI45682.1 GBXI01012495 JAD01797.1 GAKP01002678 GAKP01002677 JAC56274.1 CP012525 ALC43636.1 GANO01001671 JAB58200.1 LJIG01016003 KRT81907.1 GBYB01013759 GBYB01013761 JAG83526.1 JAG83528.1 PYGN01000143 PSN52932.1 GEZM01093979 JAV56038.1 DS235817 EEB17599.1 NNAY01003100 OXU19980.1 KQ972204 EFA12938.2 GBBI01001011 JAC17701.1 ACPB03017369 ACPB03017370 GAHY01001683 JAA75827.1 GFWV01008282 MAA33011.1 NEVH01011192 PNF32060.1 GECL01001219 JAP04905.1 GBGD01002761 JAC86128.1 GGLE01004667 MBY08793.1 GEFH01004513 JAP64068.1 GEDV01007459 JAP81098.1 GACK01002962 JAA62072.1 GBHO01044215 GBRD01005297 GDHC01014014 JAF99388.1 JAG60524.1 JAQ04615.1 GBBM01002366 JAC33052.1 GFAC01002564 JAT96624.1 AAZX01007951 GDIP01095718 JAM07997.1 GL439108 EFN67838.1 JO842912 AEO34529.1 GDIQ01081820 JAN12917.1 GDIP01233302 JAI90099.1 GDIQ01013402 JAN81335.1 GDIQ01152793 GDIQ01092666 GDIP01074909 GDIQ01031437 JAL59060.1 JAM28806.1 GL732632 EFX69694.1 KK113612 KFM60597.1 GDIQ01056879 JAN37858.1 GDIQ01152792 JAK98933.1 CCAG010000251 EZ423116 ADD19392.1 GDIQ01137151 JAL14575.1 GDIP01119661 JAL84053.1 GDIP01121654 JAL82060.1
Proteomes
UP000005204
UP000218220
UP000027135
UP000008237
UP000053825
UP000242457
+ More
UP000005203 UP000076502 UP000078542 UP000078541 UP000075809 UP000053097 UP000279307 UP000078492 UP000007755 UP000078540 UP000005205 UP000001819 UP000095300 UP000095301 UP000075840 UP000007062 UP000007798 UP000000803 UP000002282 UP000008711 UP000075880 UP000192221 UP000075883 UP000007801 UP000092461 UP000069940 UP000249989 UP000008820 UP000268350 UP000008792 UP000009192 UP000092553 UP000245037 UP000076407 UP000009046 UP000215335 UP000007266 UP000076408 UP000015103 UP000235965 UP000075884 UP000002358 UP000000311 UP000000305 UP000054359 UP000091820 UP000092443 UP000092444
UP000005203 UP000076502 UP000078542 UP000078541 UP000075809 UP000053097 UP000279307 UP000078492 UP000007755 UP000078540 UP000005205 UP000001819 UP000095300 UP000095301 UP000075840 UP000007062 UP000007798 UP000000803 UP000002282 UP000008711 UP000075880 UP000192221 UP000075883 UP000007801 UP000092461 UP000069940 UP000249989 UP000008820 UP000268350 UP000008792 UP000009192 UP000092553 UP000245037 UP000076407 UP000009046 UP000215335 UP000007266 UP000076408 UP000015103 UP000235965 UP000075884 UP000002358 UP000000311 UP000000305 UP000054359 UP000091820 UP000092443 UP000092444
Interpro
SUPFAM
SSF55811
SSF55811
ProteinModelPortal
Q1HQ96
S4P3R5
A0A1E1WIP5
A0A2W1BQ18
A0A2A4J8P7
A0A2H1WRK1
+ More
A0A067QW03 A0A1B6EAM4 E2C6H0 A0A0L7QWX6 A0A2A3EQM6 V9IF30 A0A088A0J5 A0A154P8R7 A0A195CYZ3 A0A195FWH7 A0A151WVX6 A0A026WUE0 A0A151IXD2 F4WUX3 A0A195AUB3 A0A158P0R1 A0A0R3P7I2 A0A1I8PR01 A0A1I8ML55 A0A2C9GRS2 Q7QJ32 A0A0Q9WZL1 A0A1B6MAG1 A0A0J9RSI2 Q0E8G6 A0A0R1DUQ0 A0A0Q5U3M9 A0A182JJZ8 A0A1W4VM99 A0A182MBC9 A0A1B6EQV4 A0A1B6JV95 A0A0P9C4Y2 A0A0K8TRK9 A0A1L8DGK5 A0A1B0CSL1 A0A182G2Q1 Q17DC9 A0A3B0JP98 A0A336M9B6 A0A2M4CT68 A0A2M4AXE1 A0A0Q9WIP5 A0A0Q9XMR2 A0A1Q3G177 A0A069DXT7 A0A1L8DGU6 A0A0P4VII3 A0A0K8W3P3 A0A0A1WSL8 A0A034WNR1 A0A0M5J0W6 U5EVX9 A0A0T6B375 A0A0C9S034 A0A2P8Z8W4 A0A1Y1K6G0 A0A182XLL2 E0VW43 A0A232ENQ1 D7EI10 A0A023F8J5 A0A182YMM7 R4FKV7 A0A293LYT4 A0A2J7QTZ6 A0A0V0GAF0 A0A069DPX3 A0A2R5LGY6 A0A131XD89 A0A131YR84 L7ME45 A0A0A9VUK6 A0A182NKR8 A0A023GGI1 A0A1E1XBK4 K7ISQ4 A0A0P5VTV5 E2AFE5 G3MM11 A0A0N8DXN1 A0A0P4YBY5 A0A0P6J4F1 A0A0P5S419 E9HF00 A0A087T658 A0A1A9WTL7 A0A0P6EY85 A0A0P5MVV3 A0A1A9Y0S4 D3TNW4 A0A0P5PLN6 A0A0P5V6E2 A0A0P5V2A3
A0A067QW03 A0A1B6EAM4 E2C6H0 A0A0L7QWX6 A0A2A3EQM6 V9IF30 A0A088A0J5 A0A154P8R7 A0A195CYZ3 A0A195FWH7 A0A151WVX6 A0A026WUE0 A0A151IXD2 F4WUX3 A0A195AUB3 A0A158P0R1 A0A0R3P7I2 A0A1I8PR01 A0A1I8ML55 A0A2C9GRS2 Q7QJ32 A0A0Q9WZL1 A0A1B6MAG1 A0A0J9RSI2 Q0E8G6 A0A0R1DUQ0 A0A0Q5U3M9 A0A182JJZ8 A0A1W4VM99 A0A182MBC9 A0A1B6EQV4 A0A1B6JV95 A0A0P9C4Y2 A0A0K8TRK9 A0A1L8DGK5 A0A1B0CSL1 A0A182G2Q1 Q17DC9 A0A3B0JP98 A0A336M9B6 A0A2M4CT68 A0A2M4AXE1 A0A0Q9WIP5 A0A0Q9XMR2 A0A1Q3G177 A0A069DXT7 A0A1L8DGU6 A0A0P4VII3 A0A0K8W3P3 A0A0A1WSL8 A0A034WNR1 A0A0M5J0W6 U5EVX9 A0A0T6B375 A0A0C9S034 A0A2P8Z8W4 A0A1Y1K6G0 A0A182XLL2 E0VW43 A0A232ENQ1 D7EI10 A0A023F8J5 A0A182YMM7 R4FKV7 A0A293LYT4 A0A2J7QTZ6 A0A0V0GAF0 A0A069DPX3 A0A2R5LGY6 A0A131XD89 A0A131YR84 L7ME45 A0A0A9VUK6 A0A182NKR8 A0A023GGI1 A0A1E1XBK4 K7ISQ4 A0A0P5VTV5 E2AFE5 G3MM11 A0A0N8DXN1 A0A0P4YBY5 A0A0P6J4F1 A0A0P5S419 E9HF00 A0A087T658 A0A1A9WTL7 A0A0P6EY85 A0A0P5MVV3 A0A1A9Y0S4 D3TNW4 A0A0P5PLN6 A0A0P5V6E2 A0A0P5V2A3
PDB
3N9U
E-value=7.73784e-97,
Score=899
Ontologies
GO
PANTHER
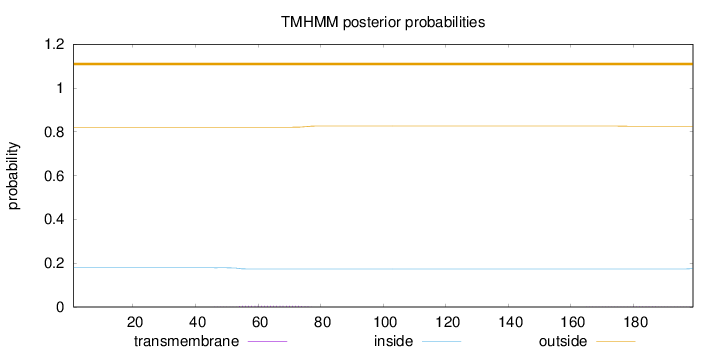
Topology
Length:
199
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14885
Exp number, first 60 AAs:
0.04025
Total prob of N-in:
0.17910
outside
1 - 199
Population Genetic Test Statistics
Pi
403.642569
Theta
225.996522
Tajima's D
2.69609
CLR
0.084021
CSRT
0.96205189740513
Interpretation
Uncertain
