Gene
KWMTBOMO06242 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001805
Annotation
PREDICTED:_28S_ribosomal_protein_S35?_mitochondrial_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 3.978
Sequence
CDS
ATGAGCATTTTAATCAAACGTTCTAAGTCTGGGTGCTTCAATAGCGCCTGCAACGTTAAATTATGGCGTCTAAACTCTACTTCGGCAGATTCCGCTCTGAAAAAGGGCGAAGAAGAAGAAGAATTTCGTGTACTAGATATCTTGAAGAAAAGGGATAAAATGCAGCGTCGAGTAGTCAAAAGAACTGATGTGCAGCCAGATAGGATCGACAGGATGGCCACAAACCAGAACTGGGGGAATGTGTGGCCTGGTCCGAAAACTTTCCACCCGTCTTCGGTGCCATTACCGATACGTCAGGGCTACGTACCAAAGGGTCAAGCACCGCCAGGTAAAAAAGCGAATGCAGAATTAATGAAAATACCAAATTTCCTTCACTTAACACCACCAGTTATCAAGAGCCAGTGTGAAGCCTTAAAACAATTTTGTACGGAATGGCCGAATCTACTCAACTCTGAGGAAAGTATAGAGAAGCATTATCCCTTAGAAATTATAAGTTCAGATTACTGTCACGCAAGTCCATCTATCCGTAACCCGTTAGCACGCGTCGTGATCTTAAGAGTTCAATTAGCAGACCTCAAACTCGATAAACATGCAACAGACAAGTTTAAGAGGCTGGTCGGTGACAGGTACGACCCAAAAACAGATTTAGTGACGATCACAGCGGATCGTTGTCCAACTAAAGTACAAAACTTGGACTATGTCAATTATTTGTTGACTGCATGCTACCATGAGTCGTGGAATTTTGAAGATTGGGAGAAGGAGAAGACCGAGGACGACATGGAGTATTATGACTGGGATAATAATGCTTCAAAGAGGAGTTTGATTGATTGGTATCTTCGTGTTAAGAATGAATTGAAAATCATGAATGATGAAGAATATAAAGCATTTGATATAAGCCAAATAGAAAATGGAGCCCAATATAAAGCGTCTGTATCAAAGTATATGAATGAAGGTGAAAATCCACAGACCGTGCAAGAATATAGTATCGCCGTTAGAAAGCTGTTTGGCTTGCCACAGAAGAAATTAGTTTAA
Protein
MSILIKRSKSGCFNSACNVKLWRLNSTSADSALKKGEEEEEFRVLDILKKRDKMQRRVVKRTDVQPDRIDRMATNQNWGNVWPGPKTFHPSSVPLPIRQGYVPKGQAPPGKKANAELMKIPNFLHLTPPVIKSQCEALKQFCTEWPNLLNSEESIEKHYPLEIISSDYCHASPSIRNPLARVVILRVQLADLKLDKHATDKFKRLVGDRYDPKTDLVTITADRCPTKVQNLDYVNYLLTACYHESWNFEDWEKEKTEDDMEYYDWDNNASKRSLIDWYLRVKNELKIMNDEEYKAFDISQIENGAQYKASVSKYMNEGENPQTVQEYSIAVRKLFGLPQKKLV
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
H9IX22
A0A2A4K395
A0A1E1WVF2
A0A2W1BK93
A0A2H1VKB2
A0A3S2LQK2
+ More
A0A0N1I9U9 A0A194Q4L8 A0A212EPG1 A0A0L7L2T0 A0A1W4XBB6 D6WDJ0 A0A0K8TT09 A0A0T6AYJ2 J3JTF0 U4U7S8 A0A232F1D7 N6T642 A0A195CBH1 A0A151X6X6 A0A195FF89 A0A158NSZ8 A0A2J7QJ66 E2A1Y3 A0A182L3Z5 A0A026WWM0 W8C7T8 Q7QIE1 A0A182IX80 A0A1B0DD95 A0A034WPA4 A0A182QF79 F4WY59 A0A182V6J2 A0A182I6D2 A0A195EHH3 A0A084WMW5 A0A182R6P4 A0A182TRX7 A0A0L7QS73 A0A182X272 A0A182MEZ2 A0A182Y890 A0A182KEN5 A0A0K8UAX5 A0A0L0CPV4 A0A1B0GJ66 A0A182PIW2 A0A336M7S7 R4G3N6 A0A0P4VMQ5 A0A023EPU6 U5EK12 A0A1Q3FBX4 E2C8E7 A0A182GF65 Q16WG3 A0A2M4AP35 A0A2M4ANQ8 A0A2M4AP49 A0A2M4BT49 A0A1S4FLQ0 A0A1Q3FCM8 A0A1Q3FEB1 A0A1L8DRJ5 A0A2M4BTG1 A0A151I467 A0A2M3Z6I5 A0A2M4BTB4 A0A2M4BTD5 W5JL09 A0A2M3ZHI2 A0A1L8DRJ8 A0A1I8MYC5 B4H1N5 A0A182WI36 Q29E49 A0A023ER01 A0A1B6KVT3 B4LE29 A0A023F7Q4 B4IYH8 A0A1A9UGC9 A0A0V0G649 A0A182N7G2 J9K252 B0WC73 Q9VZX6 A0A2M3Z6R3 B4MN34 C4WVK3 A0A224XHB3 D3TM03 A0A1A9W4C4 T1H0S3 A0A1A9YI98 A0A1Y1NCL6 A0A1I8P9U8 A0A1B0AVU1 A0A2R7W5Q1
A0A0N1I9U9 A0A194Q4L8 A0A212EPG1 A0A0L7L2T0 A0A1W4XBB6 D6WDJ0 A0A0K8TT09 A0A0T6AYJ2 J3JTF0 U4U7S8 A0A232F1D7 N6T642 A0A195CBH1 A0A151X6X6 A0A195FF89 A0A158NSZ8 A0A2J7QJ66 E2A1Y3 A0A182L3Z5 A0A026WWM0 W8C7T8 Q7QIE1 A0A182IX80 A0A1B0DD95 A0A034WPA4 A0A182QF79 F4WY59 A0A182V6J2 A0A182I6D2 A0A195EHH3 A0A084WMW5 A0A182R6P4 A0A182TRX7 A0A0L7QS73 A0A182X272 A0A182MEZ2 A0A182Y890 A0A182KEN5 A0A0K8UAX5 A0A0L0CPV4 A0A1B0GJ66 A0A182PIW2 A0A336M7S7 R4G3N6 A0A0P4VMQ5 A0A023EPU6 U5EK12 A0A1Q3FBX4 E2C8E7 A0A182GF65 Q16WG3 A0A2M4AP35 A0A2M4ANQ8 A0A2M4AP49 A0A2M4BT49 A0A1S4FLQ0 A0A1Q3FCM8 A0A1Q3FEB1 A0A1L8DRJ5 A0A2M4BTG1 A0A151I467 A0A2M3Z6I5 A0A2M4BTB4 A0A2M4BTD5 W5JL09 A0A2M3ZHI2 A0A1L8DRJ8 A0A1I8MYC5 B4H1N5 A0A182WI36 Q29E49 A0A023ER01 A0A1B6KVT3 B4LE29 A0A023F7Q4 B4IYH8 A0A1A9UGC9 A0A0V0G649 A0A182N7G2 J9K252 B0WC73 Q9VZX6 A0A2M3Z6R3 B4MN34 C4WVK3 A0A224XHB3 D3TM03 A0A1A9W4C4 T1H0S3 A0A1A9YI98 A0A1Y1NCL6 A0A1I8P9U8 A0A1B0AVU1 A0A2R7W5Q1
Pubmed
19121390
28756777
26354079
22118469
26227816
18362917
+ More
19820115 26369729 22516182 23537049 28648823 21347285 20798317 20966253 24508170 30249741 24495485 12364791 14747013 17210077 25348373 21719571 24438588 25244985 26108605 27129103 24945155 26483478 17510324 20920257 23761445 25315136 17994087 15632085 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 28004739
19820115 26369729 22516182 23537049 28648823 21347285 20798317 20966253 24508170 30249741 24495485 12364791 14747013 17210077 25348373 21719571 24438588 25244985 26108605 27129103 24945155 26483478 17510324 20920257 23761445 25315136 17994087 15632085 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 28004739
EMBL
BABH01021253
NWSH01000236
PCG78132.1
GDQN01000049
JAT91005.1
KZ149995
+ More
PZC75459.1 ODYU01003029 SOQ41289.1 RSAL01000344 RVE42340.1 KQ460038 KPJ18387.1 KQ459463 KPJ00487.1 AGBW02013491 OWR43347.1 JTDY01003285 KOB69803.1 KQ971322 EFA00783.1 GDAI01000538 JAI17065.1 LJIG01022529 KRT80098.1 BT126506 AEE61470.1 KB631768 KI210422 ERL86000.1 ERL96257.1 NNAY01001275 OXU24525.1 APGK01050964 KB741180 ENN73123.1 KQ978023 KYM98045.1 KQ982476 KYQ56050.1 KQ981636 KYN38887.1 ADTU01025462 NEVH01013559 PNF28638.1 GL435856 EFN72559.1 KK107078 QOIP01000011 EZA60475.1 RLU16865.1 GAMC01007836 GAMC01007832 GAMC01007830 GAMC01007828 JAB98719.1 AAAB01008807 EAA04180.4 AJVK01031793 GAKP01002805 JAC56147.1 AXCN02000573 GL888439 EGI60816.1 APCN01003620 KQ978957 KYN27322.1 ATLV01024521 KE525352 KFB51559.1 KQ414758 KOC61483.1 AXCM01002040 GDHF01028487 GDHF01016863 JAI23827.1 JAI35451.1 JRES01000080 KNC34378.1 AJWK01019416 UFQS01000523 UFQT01000523 SSX04566.1 SSX24929.1 ACPB03016769 GAHY01002065 JAA75445.1 GDKW01002854 JAI53741.1 GAPW01002525 JAC11073.1 GANO01001991 JAB57880.1 GFDL01009964 JAV25081.1 GL453666 EFN75782.1 JXUM01059297 KQ562045 KXJ76823.1 CH477566 EAT38934.1 GGFK01009057 MBW42378.1 GGFK01009099 MBW42420.1 GGFK01009222 MBW42543.1 GGFJ01007098 MBW56239.1 GFDL01009797 JAV25248.1 GFDL01009213 JAV25832.1 GFDF01005002 JAV09082.1 GGFJ01007153 MBW56294.1 KQ976472 KYM83990.1 GGFM01003392 MBW24143.1 GGFJ01007176 MBW56317.1 GGFJ01007178 MBW56319.1 ADMH02001223 ETN63600.1 GGFM01007253 MBW28004.1 GFDF01005001 JAV09083.1 CH479202 EDW30212.1 CH379070 EAL30213.2 GAPW01002537 JAC11061.1 GEBQ01024424 JAT15553.1 CH940647 EDW70072.1 GBBI01001311 JAC17401.1 CH916366 EDV97651.1 GECL01002600 JAP03524.1 ABLF02009803 ABLF02009804 DS231886 EDS43318.1 AE014296 BT016117 AAF47688.1 AAV37002.1 GGFM01003468 MBW24219.1 CH963847 EDW73590.1 AK341561 BAH71923.1 GFTR01004539 JAW11887.1 EZ422455 ADD18731.1 CAQQ02387843 GEZM01006628 JAV95674.1 JXJN01004388 KK854332 PTY14748.1
PZC75459.1 ODYU01003029 SOQ41289.1 RSAL01000344 RVE42340.1 KQ460038 KPJ18387.1 KQ459463 KPJ00487.1 AGBW02013491 OWR43347.1 JTDY01003285 KOB69803.1 KQ971322 EFA00783.1 GDAI01000538 JAI17065.1 LJIG01022529 KRT80098.1 BT126506 AEE61470.1 KB631768 KI210422 ERL86000.1 ERL96257.1 NNAY01001275 OXU24525.1 APGK01050964 KB741180 ENN73123.1 KQ978023 KYM98045.1 KQ982476 KYQ56050.1 KQ981636 KYN38887.1 ADTU01025462 NEVH01013559 PNF28638.1 GL435856 EFN72559.1 KK107078 QOIP01000011 EZA60475.1 RLU16865.1 GAMC01007836 GAMC01007832 GAMC01007830 GAMC01007828 JAB98719.1 AAAB01008807 EAA04180.4 AJVK01031793 GAKP01002805 JAC56147.1 AXCN02000573 GL888439 EGI60816.1 APCN01003620 KQ978957 KYN27322.1 ATLV01024521 KE525352 KFB51559.1 KQ414758 KOC61483.1 AXCM01002040 GDHF01028487 GDHF01016863 JAI23827.1 JAI35451.1 JRES01000080 KNC34378.1 AJWK01019416 UFQS01000523 UFQT01000523 SSX04566.1 SSX24929.1 ACPB03016769 GAHY01002065 JAA75445.1 GDKW01002854 JAI53741.1 GAPW01002525 JAC11073.1 GANO01001991 JAB57880.1 GFDL01009964 JAV25081.1 GL453666 EFN75782.1 JXUM01059297 KQ562045 KXJ76823.1 CH477566 EAT38934.1 GGFK01009057 MBW42378.1 GGFK01009099 MBW42420.1 GGFK01009222 MBW42543.1 GGFJ01007098 MBW56239.1 GFDL01009797 JAV25248.1 GFDL01009213 JAV25832.1 GFDF01005002 JAV09082.1 GGFJ01007153 MBW56294.1 KQ976472 KYM83990.1 GGFM01003392 MBW24143.1 GGFJ01007176 MBW56317.1 GGFJ01007178 MBW56319.1 ADMH02001223 ETN63600.1 GGFM01007253 MBW28004.1 GFDF01005001 JAV09083.1 CH479202 EDW30212.1 CH379070 EAL30213.2 GAPW01002537 JAC11061.1 GEBQ01024424 JAT15553.1 CH940647 EDW70072.1 GBBI01001311 JAC17401.1 CH916366 EDV97651.1 GECL01002600 JAP03524.1 ABLF02009803 ABLF02009804 DS231886 EDS43318.1 AE014296 BT016117 AAF47688.1 AAV37002.1 GGFM01003468 MBW24219.1 CH963847 EDW73590.1 AK341561 BAH71923.1 GFTR01004539 JAW11887.1 EZ422455 ADD18731.1 CAQQ02387843 GEZM01006628 JAV95674.1 JXJN01004388 KK854332 PTY14748.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000037510 UP000192223 UP000007266 UP000030742 UP000215335 UP000019118 UP000078542 UP000075809 UP000078541 UP000005205 UP000235965 UP000000311 UP000075882 UP000053097 UP000279307 UP000007062 UP000075880 UP000092462 UP000075886 UP000007755 UP000075903 UP000075840 UP000078492 UP000030765 UP000075900 UP000075902 UP000053825 UP000076407 UP000075883 UP000076408 UP000075881 UP000037069 UP000092461 UP000075885 UP000015103 UP000008237 UP000069940 UP000249989 UP000008820 UP000078540 UP000000673 UP000095301 UP000008744 UP000075920 UP000001819 UP000008792 UP000001070 UP000078200 UP000075884 UP000007819 UP000002320 UP000000803 UP000007798 UP000091820 UP000015102 UP000092443 UP000095300 UP000092460
UP000037510 UP000192223 UP000007266 UP000030742 UP000215335 UP000019118 UP000078542 UP000075809 UP000078541 UP000005205 UP000235965 UP000000311 UP000075882 UP000053097 UP000279307 UP000007062 UP000075880 UP000092462 UP000075886 UP000007755 UP000075903 UP000075840 UP000078492 UP000030765 UP000075900 UP000075902 UP000053825 UP000076407 UP000075883 UP000076408 UP000075881 UP000037069 UP000092461 UP000075885 UP000015103 UP000008237 UP000069940 UP000249989 UP000008820 UP000078540 UP000000673 UP000095301 UP000008744 UP000075920 UP000001819 UP000008792 UP000001070 UP000078200 UP000075884 UP000007819 UP000002320 UP000000803 UP000007798 UP000091820 UP000015102 UP000092443 UP000095300 UP000092460
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
H9IX22
A0A2A4K395
A0A1E1WVF2
A0A2W1BK93
A0A2H1VKB2
A0A3S2LQK2
+ More
A0A0N1I9U9 A0A194Q4L8 A0A212EPG1 A0A0L7L2T0 A0A1W4XBB6 D6WDJ0 A0A0K8TT09 A0A0T6AYJ2 J3JTF0 U4U7S8 A0A232F1D7 N6T642 A0A195CBH1 A0A151X6X6 A0A195FF89 A0A158NSZ8 A0A2J7QJ66 E2A1Y3 A0A182L3Z5 A0A026WWM0 W8C7T8 Q7QIE1 A0A182IX80 A0A1B0DD95 A0A034WPA4 A0A182QF79 F4WY59 A0A182V6J2 A0A182I6D2 A0A195EHH3 A0A084WMW5 A0A182R6P4 A0A182TRX7 A0A0L7QS73 A0A182X272 A0A182MEZ2 A0A182Y890 A0A182KEN5 A0A0K8UAX5 A0A0L0CPV4 A0A1B0GJ66 A0A182PIW2 A0A336M7S7 R4G3N6 A0A0P4VMQ5 A0A023EPU6 U5EK12 A0A1Q3FBX4 E2C8E7 A0A182GF65 Q16WG3 A0A2M4AP35 A0A2M4ANQ8 A0A2M4AP49 A0A2M4BT49 A0A1S4FLQ0 A0A1Q3FCM8 A0A1Q3FEB1 A0A1L8DRJ5 A0A2M4BTG1 A0A151I467 A0A2M3Z6I5 A0A2M4BTB4 A0A2M4BTD5 W5JL09 A0A2M3ZHI2 A0A1L8DRJ8 A0A1I8MYC5 B4H1N5 A0A182WI36 Q29E49 A0A023ER01 A0A1B6KVT3 B4LE29 A0A023F7Q4 B4IYH8 A0A1A9UGC9 A0A0V0G649 A0A182N7G2 J9K252 B0WC73 Q9VZX6 A0A2M3Z6R3 B4MN34 C4WVK3 A0A224XHB3 D3TM03 A0A1A9W4C4 T1H0S3 A0A1A9YI98 A0A1Y1NCL6 A0A1I8P9U8 A0A1B0AVU1 A0A2R7W5Q1
A0A0N1I9U9 A0A194Q4L8 A0A212EPG1 A0A0L7L2T0 A0A1W4XBB6 D6WDJ0 A0A0K8TT09 A0A0T6AYJ2 J3JTF0 U4U7S8 A0A232F1D7 N6T642 A0A195CBH1 A0A151X6X6 A0A195FF89 A0A158NSZ8 A0A2J7QJ66 E2A1Y3 A0A182L3Z5 A0A026WWM0 W8C7T8 Q7QIE1 A0A182IX80 A0A1B0DD95 A0A034WPA4 A0A182QF79 F4WY59 A0A182V6J2 A0A182I6D2 A0A195EHH3 A0A084WMW5 A0A182R6P4 A0A182TRX7 A0A0L7QS73 A0A182X272 A0A182MEZ2 A0A182Y890 A0A182KEN5 A0A0K8UAX5 A0A0L0CPV4 A0A1B0GJ66 A0A182PIW2 A0A336M7S7 R4G3N6 A0A0P4VMQ5 A0A023EPU6 U5EK12 A0A1Q3FBX4 E2C8E7 A0A182GF65 Q16WG3 A0A2M4AP35 A0A2M4ANQ8 A0A2M4AP49 A0A2M4BT49 A0A1S4FLQ0 A0A1Q3FCM8 A0A1Q3FEB1 A0A1L8DRJ5 A0A2M4BTG1 A0A151I467 A0A2M3Z6I5 A0A2M4BTB4 A0A2M4BTD5 W5JL09 A0A2M3ZHI2 A0A1L8DRJ8 A0A1I8MYC5 B4H1N5 A0A182WI36 Q29E49 A0A023ER01 A0A1B6KVT3 B4LE29 A0A023F7Q4 B4IYH8 A0A1A9UGC9 A0A0V0G649 A0A182N7G2 J9K252 B0WC73 Q9VZX6 A0A2M3Z6R3 B4MN34 C4WVK3 A0A224XHB3 D3TM03 A0A1A9W4C4 T1H0S3 A0A1A9YI98 A0A1Y1NCL6 A0A1I8P9U8 A0A1B0AVU1 A0A2R7W5Q1
PDB
6GAZ
E-value=1.10464e-72,
Score=694
Ontologies
GO
Topology
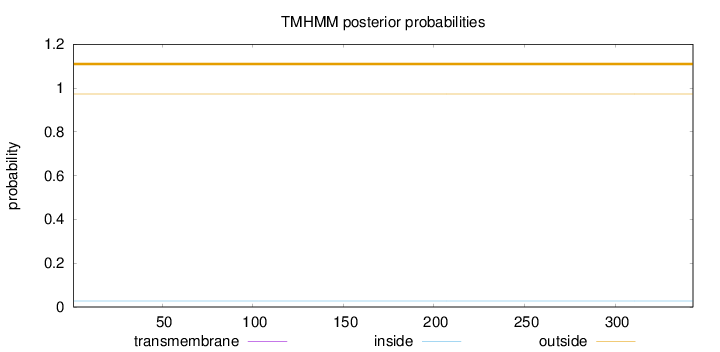
Length:
343
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00014
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02754
outside
1 - 343
Population Genetic Test Statistics
Pi
235.608935
Theta
198.735547
Tajima's D
0.540846
CLR
1.291843
CSRT
0.528723563821809
Interpretation
Uncertain
