Gene
KWMTBOMO06235 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001638
Annotation
PREDICTED:_xaa-Pro_aminopeptidase_1_[Bombyx_mori]
Full name
Xaa-Pro aminopeptidase ApepP
Alternative Name
Aminopeptidase P
Location in the cell
Cytoplasmic Reliability : 1.625 Mitochondrial Reliability : 1.61
Sequence
CDS
ATGCTAGCCGTCGCACTGATCGTCATTTTAACAGTCAAATGTAGCCTGACAGAAGGCGCTTTGGCCAGTAACAGTTCTATCGACCGAAAAGATCGCAACCGCGAGATGTCGCTACAGCGCTTGCAAGCGCTCCGAGCGCTAATGTCGAGTCAGCGTCCCGTGTTGGCAGCTTATATTATACCGACCGCCGATGCTCATAATTCGGAGTACATAGATGCAGCGGACGCCAGACGCGAGTGGATATCCGCCTTCACCGGGTCGGCTGGCACTGCAGTTGTGACGTCATCGCAGGCTCTCGTCTGGACCGACGGTAGATACTACACGCAGTTCGAGAGGGAGGTCGACTTGTCAGCGTGGACTTTAATGAAACAGACACTACCGGACACTCCCACTTTGGAGAAATGGCTGGCCTCGAACCTCAAAGACGGTGACGTAGTCGGCGTGGATCCCCAAACGATGACCAGAGACGAATGGACTCCGATACAGACTGCCCTAAAGAAAATTAGCGCTCAACTAGTGCCGATTTCGAACAACCTCGTGGATGATGTCAGGATTGAACTTGGGGACCCGGCGCCTAAGAGATCGCACAATGAACTCGCGCCATTACACGTCCGCTACACTGGCAGAACAGCGGGAGAGAAGATCGCGGAATTAAGACGGAAGATGGCGGAGAAGAAAGCTTCGGCCCTCGTTCTAACGGCCTTAGACGATATTGCTTACACGTTAAATCTACGCGGCTCCGACATCGAATATAATCCGGTCTTCTTTTCGTACCTGATCGTGATGGAGACCAATGTTGTTCTATACTGGGGCGACGGTCAACTGCCCCCCTCCGTGACGGATCACCTGAGGGAGGAGGGCGTGGACAGTGTCGAGGGCAAACCTTACGGGGACATTTTGGAGGGCTTAAAGGAAATGGCGCGAGAACTGTCGGAGAGCGGCGACGGGAGACACGTGATCTGGTTGTCCAATGACGCTAATGAGGCGATCCACAGAGCCGCCTCCGGGTCTGATGTCCTAAAGCGTCCGATTGATTTAATATCGGAGGTGTCGCCCGTGAAACTGGCTAAATTGGTCAAAAACGAAATCGAACTGCAAGGCTTCCGTAACTGCCACATCAAGGACGGTATAGCCGTGGTCAGGTTCTTCAGGTGGCTCCACGATCAAATTGACAGCGGCGAGCAGATCACAGAGGTGCAGGCTTCGGATAAACTTCTGGAGTTTAGGAGGGACGAAAAGGACTTCATGGGTCCTTCGTTTGGAACGATAGCGGGCAGCGGCGCGAACGGCGCAATCATACACTACAGCCCCGATAGAGAGGGACAGCAGACCGTGATAGAGAGAGACCACATGTTCCTTCTCGACTCCGGCGGGCAATACAGGGATGGCACGACCGACATAACTCGCACGCGGCACATGAGCGGCAACCCTACCGCGGAACAGAAGGATGCGTTCACTCGAGTGCTAAAAGGTCAGATTATGATCGGCAGCGCTTTATTCCCGAAAGGCGTTAAGGGCAACGTTCTGGACAGCTTCGCGCGTCACGCCCTGTGGGAGGTGGGCCTGGACTACGGTCACGGGACGGGTCACGGCGTGGGTCACTACCTCAACGTGCACGAGGGTCCGTCCGGAGTCTCTTGGCGTCCGTACCCCGACGATCCTGGACTGAACGTTGGACAGATATTGAGCAACGAACCAGGCTTCTACAAAGTCGGCGAATACGGAATTCGCCACGAAGATCTAATAGAGATTGTGGCTGTCGATAAGGGAAGCGATCATCCTAAGGCGAAGAATCTGAGAGGCGATTACGATGGTCGAGGCGTTTTGGGCTTCAGCACGTTGACTTTAGTGCCCAATCAAAGGGAATGTATCGATGCAGAGATTATGACCGATTTCGAGATTTCATACGTGAACGCGTACCATCGAAGAGTGCTTGACACATTGAGTCCGATACTGAAGGAGCGCGGTTTGCTTAAGGATTTGGAGTGGCTCGAAGGTCAATGCATCCCGATAACCAGGAAGTGA
Protein
MLAVALIVILTVKCSLTEGALASNSSIDRKDRNREMSLQRLQALRALMSSQRPVLAAYIIPTADAHNSEYIDAADARREWISAFTGSAGTAVVTSSQALVWTDGRYYTQFEREVDLSAWTLMKQTLPDTPTLEKWLASNLKDGDVVGVDPQTMTRDEWTPIQTALKKISAQLVPISNNLVDDVRIELGDPAPKRSHNELAPLHVRYTGRTAGEKIAELRRKMAEKKASALVLTALDDIAYTLNLRGSDIEYNPVFFSYLIVMETNVVLYWGDGQLPPSVTDHLREEGVDSVEGKPYGDILEGLKEMARELSESGDGRHVIWLSNDANEAIHRAASGSDVLKRPIDLISEVSPVKLAKLVKNEIELQGFRNCHIKDGIAVVRFFRWLHDQIDSGEQITEVQASDKLLEFRRDEKDFMGPSFGTIAGSGANGAIIHYSPDREGQQTVIERDHMFLLDSGGQYRDGTTDITRTRHMSGNPTAEQKDAFTRVLKGQIMIGSALFPKGVKGNVLDSFARHALWEVGLDYGHGTGHGVGHYLNVHEGPSGVSWRPYPDDPGLNVGQILSNEPGFYKVGEYGIRHEDLIEIVAVDKGSDHPKAKNLRGDYDGRGVLGFSTLTLVPNQRECIDAEIMTDFEISYVNAYHRRVLDTLSPILKERGLLKDLEWLEGQCIPITRK
Summary
Description
Catalyzes the removal of a penultimate prolyl residue from the N-termini of peptides, such as Arg-Pro-Pro.
Catalytic Activity
Release of any N-terminal amino acid, including proline, that is linked to proline, even from a dipeptide or tripeptide.
Cofactor
Mn(2+)
Biophysicochemical Properties
Optimum pH is 7.6.
Similarity
Belongs to the peptidase M24B family.
Keywords
Aminopeptidase
Complete proteome
Cytoplasm
Hydrolase
Manganese
Metal-binding
Protease
Reference proteome
Feature
chain Xaa-Pro aminopeptidase ApepP
Uniprot
H9IWK6
A0A2A4K1N0
A0A194Q542
A0A0N0PEM0
A0A194Q663
F2YHL8
+ More
A0A2H1WE50 A0A2A4JIL6 A0A0T6ATZ3 A0A182JDI6 A0A182N8Z8 A0A0A1XP05 A0A182W4V1 D6WLY7 A0A182Y514 A0A182Q0F4 A0A1W4VXW7 A0A182M9W8 A0A1L8EFL3 A0A1L8EFY8 A0A1L8EFT0 A0A182XPM7 A0A1L8EFQ1 A0A182PNQ4 A0A182RP79 A0A1L8EFL9 A0A034UZ61 Q5TW92 A0A182VC76 A0A182K5H5 A0A182KL97 A0A0K8VUN3 A0A2M3Z589 A0A1Q3G343 A0A182IDV1 A0A0K8TT40 A0A182FME0 A0A1S4FNP6 Q16UX2 A0A1S4FNB2 Q16UX1 A0A1I8NYB9 T1DPD1 A0A2M4BHK4 A0A1I8NYD3 B5DWN6 A0A1J1INB7 A0A2M4BHM1 A0A1Y1MRV6 B3MML0 A0A023EVH9 W5JQL7 A0A3B0KAY1 B4GE57 A0A2M4AFN2 W8C8S2 A0A2M4AFQ4 O96794 Q9VJG0 B4NBP8 E0V912 A0A0J9R3N8 A0A0P5N2B9 A0A0P5YIN6 A0A0P5UYA9 B4P8U5 A0A0P5AQ29 B3NLF8 A0A0P6AUE6 A0A0P6CIW7 A0A0P5ZQD5 A0A0P6A1X3 B4Q7I7 A0A0P4ZJN6 A0A1L8E2Y5 A0A1L8E2K1 A0A0P5V5V5 A0A0P5S2U6 A0A0P5MR17 A0A0P5JZH2 A0A0P6GIR2 A0A0P4XLX8 A0A0P5MKE7 A0A0P6CY66 A0A0P5VVJ4 T1PD98 A0A0P4XT88 A0A1A9YT01 A0A1A9WYM0 A0A0P5HT27 B4L176 A0A1A9VFT0 R4FNJ0 A0A067RCX1 A0A1I8N275 A0A0P4VTP9 A0A2J7PRP2
A0A2H1WE50 A0A2A4JIL6 A0A0T6ATZ3 A0A182JDI6 A0A182N8Z8 A0A0A1XP05 A0A182W4V1 D6WLY7 A0A182Y514 A0A182Q0F4 A0A1W4VXW7 A0A182M9W8 A0A1L8EFL3 A0A1L8EFY8 A0A1L8EFT0 A0A182XPM7 A0A1L8EFQ1 A0A182PNQ4 A0A182RP79 A0A1L8EFL9 A0A034UZ61 Q5TW92 A0A182VC76 A0A182K5H5 A0A182KL97 A0A0K8VUN3 A0A2M3Z589 A0A1Q3G343 A0A182IDV1 A0A0K8TT40 A0A182FME0 A0A1S4FNP6 Q16UX2 A0A1S4FNB2 Q16UX1 A0A1I8NYB9 T1DPD1 A0A2M4BHK4 A0A1I8NYD3 B5DWN6 A0A1J1INB7 A0A2M4BHM1 A0A1Y1MRV6 B3MML0 A0A023EVH9 W5JQL7 A0A3B0KAY1 B4GE57 A0A2M4AFN2 W8C8S2 A0A2M4AFQ4 O96794 Q9VJG0 B4NBP8 E0V912 A0A0J9R3N8 A0A0P5N2B9 A0A0P5YIN6 A0A0P5UYA9 B4P8U5 A0A0P5AQ29 B3NLF8 A0A0P6AUE6 A0A0P6CIW7 A0A0P5ZQD5 A0A0P6A1X3 B4Q7I7 A0A0P4ZJN6 A0A1L8E2Y5 A0A1L8E2K1 A0A0P5V5V5 A0A0P5S2U6 A0A0P5MR17 A0A0P5JZH2 A0A0P6GIR2 A0A0P4XLX8 A0A0P5MKE7 A0A0P6CY66 A0A0P5VVJ4 T1PD98 A0A0P4XT88 A0A1A9YT01 A0A1A9WYM0 A0A0P5HT27 B4L176 A0A1A9VFT0 R4FNJ0 A0A067RCX1 A0A1I8N275 A0A0P4VTP9 A0A2J7PRP2
EC Number
3.4.11.9
Pubmed
EMBL
BABH01021246
NWSH01000280
PCG77804.1
KQ459463
KPJ00479.1
KQ459920
+ More
KPJ19323.1 KPJ00495.1 JF262040 ADZ74177.1 ODYU01008066 SOQ51348.1 NWSH01001426 PCG71273.1 LJIG01022812 KRT78593.1 GBXI01001615 JAD12677.1 KQ971343 EFA04188.1 AXCN02001401 AXCM01002320 GFDG01001277 JAV17522.1 GFDG01001280 JAV17519.1 GFDG01001283 JAV17516.1 GFDG01001281 JAV17518.1 GFDG01001282 JAV17517.1 GAKP01022691 JAC36261.1 AAAB01008811 EAL41692.4 GDHF01009720 JAI42594.1 GGFM01002936 MBW23687.1 GFDL01000869 JAV34176.1 APCN01004825 GDAI01000081 JAI17522.1 CH477612 EAT38334.1 EAT38333.1 GAMD01003444 JAA98146.1 GGFJ01003395 MBW52536.1 CM000070 EDY68177.2 CVRI01000054 CRL00590.1 GGFJ01003396 MBW52537.1 GEZM01029634 JAV86027.1 CH902620 EDV30956.1 GAPW01000577 JAC13021.1 ADMH02000792 ETN65044.1 OUUW01000007 SPP83259.1 CH479182 EDW33892.1 GGFK01006280 MBW39601.1 GAMC01002794 JAC03762.1 GGFK01006293 MBW39614.1 AJ131920 CAA10526.2 AY082012 AE014134 AY061297 AAL28845.1 AAL99293.1 CH964232 EDW81212.2 DS234988 EEB09868.1 CM002910 KMY90641.1 GDIQ01148334 JAL03392.1 GDIP01058753 JAM44962.1 GDIP01107138 JAL96576.1 CM000158 EDW90203.1 GDIP01196076 GDIP01081431 JAJ27326.1 CH954179 EDV54874.1 GDIP01038153 JAM65562.1 GDIQ01090633 JAN04104.1 GDIP01041094 JAM62621.1 GDIP01040200 JAM63515.1 CM000361 EDX05285.1 GDIP01215680 JAJ07722.1 GFDF01001142 JAV12942.1 GFDF01001143 JAV12941.1 GDIP01104536 JAL99178.1 GDIQ01145525 GDIQ01093087 GDIQ01030273 JAL58639.1 GDIQ01152570 JAK99155.1 GDIQ01191342 GDIQ01036630 JAK60383.1 GDIQ01108886 GDIQ01052418 JAN42319.1 GDIP01239376 JAI84025.1 GDIQ01155365 JAK96360.1 GDIQ01088145 GDIQ01086396 JAN06592.1 GDIP01094597 JAM09118.1 KA646762 AFP61391.1 GDIP01238077 JAI85324.1 GDIQ01229144 JAK22581.1 CH933809 EDW19258.2 ACPB03006450 GAHY01001269 JAA76241.1 KK852543 KDR21696.1 GDKW01001241 JAI55354.1 NEVH01021955 PNF19004.1
KPJ19323.1 KPJ00495.1 JF262040 ADZ74177.1 ODYU01008066 SOQ51348.1 NWSH01001426 PCG71273.1 LJIG01022812 KRT78593.1 GBXI01001615 JAD12677.1 KQ971343 EFA04188.1 AXCN02001401 AXCM01002320 GFDG01001277 JAV17522.1 GFDG01001280 JAV17519.1 GFDG01001283 JAV17516.1 GFDG01001281 JAV17518.1 GFDG01001282 JAV17517.1 GAKP01022691 JAC36261.1 AAAB01008811 EAL41692.4 GDHF01009720 JAI42594.1 GGFM01002936 MBW23687.1 GFDL01000869 JAV34176.1 APCN01004825 GDAI01000081 JAI17522.1 CH477612 EAT38334.1 EAT38333.1 GAMD01003444 JAA98146.1 GGFJ01003395 MBW52536.1 CM000070 EDY68177.2 CVRI01000054 CRL00590.1 GGFJ01003396 MBW52537.1 GEZM01029634 JAV86027.1 CH902620 EDV30956.1 GAPW01000577 JAC13021.1 ADMH02000792 ETN65044.1 OUUW01000007 SPP83259.1 CH479182 EDW33892.1 GGFK01006280 MBW39601.1 GAMC01002794 JAC03762.1 GGFK01006293 MBW39614.1 AJ131920 CAA10526.2 AY082012 AE014134 AY061297 AAL28845.1 AAL99293.1 CH964232 EDW81212.2 DS234988 EEB09868.1 CM002910 KMY90641.1 GDIQ01148334 JAL03392.1 GDIP01058753 JAM44962.1 GDIP01107138 JAL96576.1 CM000158 EDW90203.1 GDIP01196076 GDIP01081431 JAJ27326.1 CH954179 EDV54874.1 GDIP01038153 JAM65562.1 GDIQ01090633 JAN04104.1 GDIP01041094 JAM62621.1 GDIP01040200 JAM63515.1 CM000361 EDX05285.1 GDIP01215680 JAJ07722.1 GFDF01001142 JAV12942.1 GFDF01001143 JAV12941.1 GDIP01104536 JAL99178.1 GDIQ01145525 GDIQ01093087 GDIQ01030273 JAL58639.1 GDIQ01152570 JAK99155.1 GDIQ01191342 GDIQ01036630 JAK60383.1 GDIQ01108886 GDIQ01052418 JAN42319.1 GDIP01239376 JAI84025.1 GDIQ01155365 JAK96360.1 GDIQ01088145 GDIQ01086396 JAN06592.1 GDIP01094597 JAM09118.1 KA646762 AFP61391.1 GDIP01238077 JAI85324.1 GDIQ01229144 JAK22581.1 CH933809 EDW19258.2 ACPB03006450 GAHY01001269 JAA76241.1 KK852543 KDR21696.1 GDKW01001241 JAI55354.1 NEVH01021955 PNF19004.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000075880
UP000075884
+ More
UP000075920 UP000007266 UP000076408 UP000075886 UP000192221 UP000075883 UP000076407 UP000075885 UP000075900 UP000007062 UP000075903 UP000075881 UP000075882 UP000075840 UP000069272 UP000008820 UP000095300 UP000001819 UP000183832 UP000007801 UP000000673 UP000268350 UP000008744 UP000000803 UP000007798 UP000009046 UP000002282 UP000008711 UP000000304 UP000092443 UP000091820 UP000009192 UP000078200 UP000015103 UP000027135 UP000095301 UP000235965
UP000075920 UP000007266 UP000076408 UP000075886 UP000192221 UP000075883 UP000076407 UP000075885 UP000075900 UP000007062 UP000075903 UP000075881 UP000075882 UP000075840 UP000069272 UP000008820 UP000095300 UP000001819 UP000183832 UP000007801 UP000000673 UP000268350 UP000008744 UP000000803 UP000007798 UP000009046 UP000002282 UP000008711 UP000000304 UP000092443 UP000091820 UP000009192 UP000078200 UP000015103 UP000027135 UP000095301 UP000235965
Interpro
Gene 3D
CDD
ProteinModelPortal
H9IWK6
A0A2A4K1N0
A0A194Q542
A0A0N0PEM0
A0A194Q663
F2YHL8
+ More
A0A2H1WE50 A0A2A4JIL6 A0A0T6ATZ3 A0A182JDI6 A0A182N8Z8 A0A0A1XP05 A0A182W4V1 D6WLY7 A0A182Y514 A0A182Q0F4 A0A1W4VXW7 A0A182M9W8 A0A1L8EFL3 A0A1L8EFY8 A0A1L8EFT0 A0A182XPM7 A0A1L8EFQ1 A0A182PNQ4 A0A182RP79 A0A1L8EFL9 A0A034UZ61 Q5TW92 A0A182VC76 A0A182K5H5 A0A182KL97 A0A0K8VUN3 A0A2M3Z589 A0A1Q3G343 A0A182IDV1 A0A0K8TT40 A0A182FME0 A0A1S4FNP6 Q16UX2 A0A1S4FNB2 Q16UX1 A0A1I8NYB9 T1DPD1 A0A2M4BHK4 A0A1I8NYD3 B5DWN6 A0A1J1INB7 A0A2M4BHM1 A0A1Y1MRV6 B3MML0 A0A023EVH9 W5JQL7 A0A3B0KAY1 B4GE57 A0A2M4AFN2 W8C8S2 A0A2M4AFQ4 O96794 Q9VJG0 B4NBP8 E0V912 A0A0J9R3N8 A0A0P5N2B9 A0A0P5YIN6 A0A0P5UYA9 B4P8U5 A0A0P5AQ29 B3NLF8 A0A0P6AUE6 A0A0P6CIW7 A0A0P5ZQD5 A0A0P6A1X3 B4Q7I7 A0A0P4ZJN6 A0A1L8E2Y5 A0A1L8E2K1 A0A0P5V5V5 A0A0P5S2U6 A0A0P5MR17 A0A0P5JZH2 A0A0P6GIR2 A0A0P4XLX8 A0A0P5MKE7 A0A0P6CY66 A0A0P5VVJ4 T1PD98 A0A0P4XT88 A0A1A9YT01 A0A1A9WYM0 A0A0P5HT27 B4L176 A0A1A9VFT0 R4FNJ0 A0A067RCX1 A0A1I8N275 A0A0P4VTP9 A0A2J7PRP2
A0A2H1WE50 A0A2A4JIL6 A0A0T6ATZ3 A0A182JDI6 A0A182N8Z8 A0A0A1XP05 A0A182W4V1 D6WLY7 A0A182Y514 A0A182Q0F4 A0A1W4VXW7 A0A182M9W8 A0A1L8EFL3 A0A1L8EFY8 A0A1L8EFT0 A0A182XPM7 A0A1L8EFQ1 A0A182PNQ4 A0A182RP79 A0A1L8EFL9 A0A034UZ61 Q5TW92 A0A182VC76 A0A182K5H5 A0A182KL97 A0A0K8VUN3 A0A2M3Z589 A0A1Q3G343 A0A182IDV1 A0A0K8TT40 A0A182FME0 A0A1S4FNP6 Q16UX2 A0A1S4FNB2 Q16UX1 A0A1I8NYB9 T1DPD1 A0A2M4BHK4 A0A1I8NYD3 B5DWN6 A0A1J1INB7 A0A2M4BHM1 A0A1Y1MRV6 B3MML0 A0A023EVH9 W5JQL7 A0A3B0KAY1 B4GE57 A0A2M4AFN2 W8C8S2 A0A2M4AFQ4 O96794 Q9VJG0 B4NBP8 E0V912 A0A0J9R3N8 A0A0P5N2B9 A0A0P5YIN6 A0A0P5UYA9 B4P8U5 A0A0P5AQ29 B3NLF8 A0A0P6AUE6 A0A0P6CIW7 A0A0P5ZQD5 A0A0P6A1X3 B4Q7I7 A0A0P4ZJN6 A0A1L8E2Y5 A0A1L8E2K1 A0A0P5V5V5 A0A0P5S2U6 A0A0P5MR17 A0A0P5JZH2 A0A0P6GIR2 A0A0P4XLX8 A0A0P5MKE7 A0A0P6CY66 A0A0P5VVJ4 T1PD98 A0A0P4XT88 A0A1A9YT01 A0A1A9WYM0 A0A0P5HT27 B4L176 A0A1A9VFT0 R4FNJ0 A0A067RCX1 A0A1I8N275 A0A0P4VTP9 A0A2J7PRP2
PDB
3CTZ
E-value=1.16215e-133,
Score=1223
Ontologies
GO
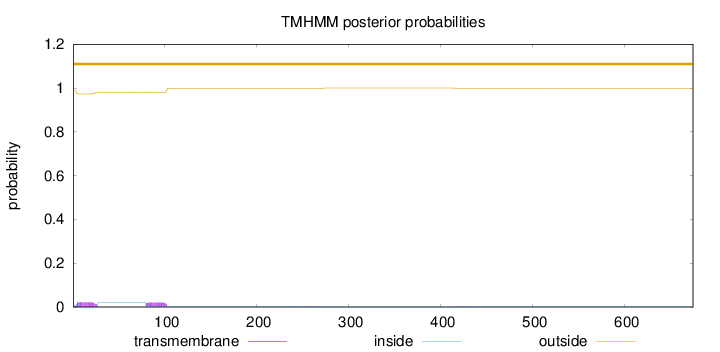
Topology
Subcellular location
Cytoplasm
SignalP
Position: 1 - 19,
Likelihood: 0.752820
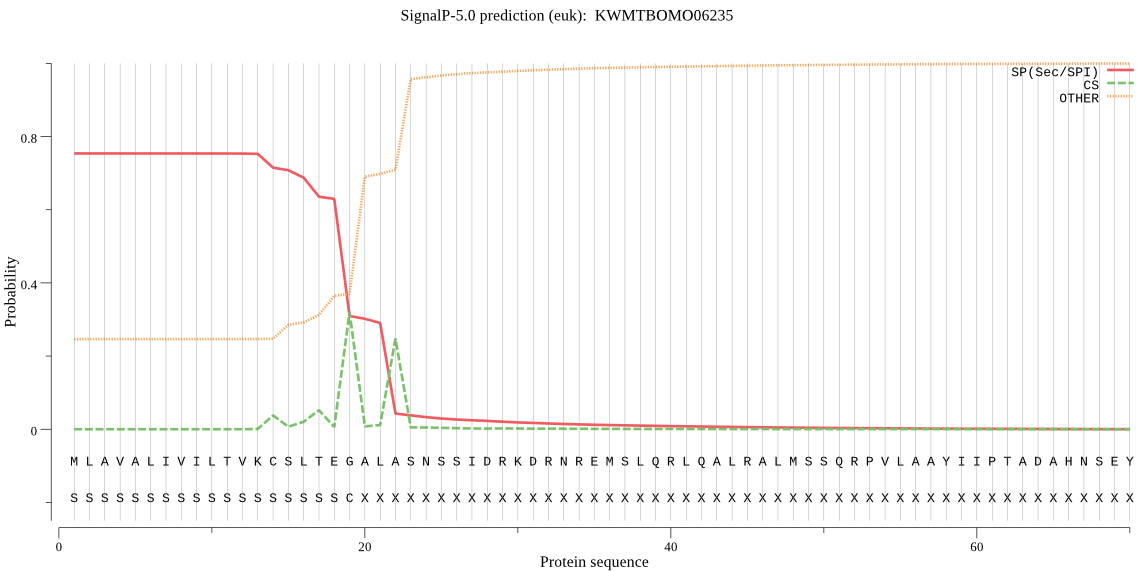
Length:
674
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.9563
Exp number, first 60 AAs:
0.47144
Total prob of N-in:
0.01115
outside
1 - 674
Population Genetic Test Statistics
Pi
257.893549
Theta
218.959086
Tajima's D
1.750412
CLR
0.120774
CSRT
0.84110794460277
Interpretation
Uncertain
