Pre Gene Modal
BGIBMGA001801
Annotation
Uncharacterized_protein_OBRU01_01903_[Operophtera_brumata]
Full name
RuvB-like helicase
Location in the cell
Cytoplasmic Reliability : 1.329 Nuclear Reliability : 1.543
Sequence
CDS
ATGGAAGACGTGAAAAAGGTCTACTCGCTGTTCTTGGACGAGCATCGTTCCGAGCAGTTTTTGAAGGAGTACCAGGACGAGTTTATGTTTAACGATGGTTCCGGCGATGCATCGCAAGTGATGATGGAAGTATCTCAAAATTTACTTCGCGCTAACAACCAAATCTTTAAGAATGTCGCCCAACAAAGGAACATGAGCGTTATCTGCACTCCGCCAAGAAACAAGGTTTCCAGAGGTGAGATGATCTTCCTCGCCGGCCTGATGGTGGTGGGCTGGTCTGCCATCCCAGCCTGGGTGTTGGTCAATATCAAGCATTACCGCGACAAGCAATAA
Protein
MEDVKKVYSLFLDEHRSEQFLKEYQDEFMFNDGSGDASQVMMEVSQNLLRANNQIFKNVAQQRNMSVICTPPRNKVSRGEMIFLAGLMVVGWSAIPAWVLVNIKHYRDKQ
Summary
Description
Proposed core component of the chromatin remodeling INO80 complex which is involved in transcriptional regulation, DNA replication and probably DNA repair.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Similarity
Belongs to the RuvB family.
Uniprot
EC Number
3.6.4.12
Pubmed
EMBL
Proteomes
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
PDB
3UK6
E-value=4.68517e-09,
Score=139
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Nucleus
Length:
110
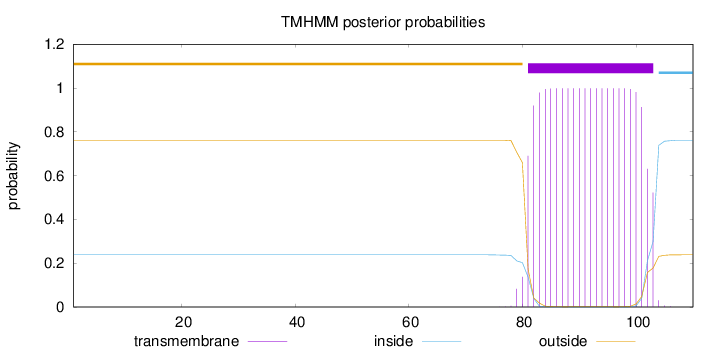
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.87819
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.23811
outside
1 - 80
TMhelix
81 - 103
inside
104 - 110
Population Genetic Test Statistics
Pi
222.255046
Theta
143.266381
Tajima's D
0.855103
CLR
10.01176
CSRT
0.621568921553922
Interpretation
Uncertain
