Pre Gene Modal
BGIBMGA001801
Annotation
PREDICTED:_ruvB-like_2_[Papilio_polytes]
Full name
RuvB-like helicase
+ More
RuvB-like 2
RuvB-like 2
Alternative Name
Reptin
p47 protein
p47 protein
Location in the cell
Mitochondrial Reliability : 2.129
Sequence
CDS
ATGGCTTCAATAGCAGCGGCGCAAGTACAAGAAGTGCGTTCTATAACGCGCATCGAGAGAATTGGCGCGCATTCCCACATTAGAGGACTGGGCTTAGATGACTCTTTGGAGCCAAGACAAGTATCGCAGGGCATGGTCGGCCAGAAGATGGCAAGGAAGGCCGCCGGAGTCATCTTACAGATGATTCGAGAAGGTAAGATAGCGGGACGAGCAGTGTTATTGGCCGGTCAGCCTGGTACAGGGAAGACTGCCATCGCTATGGGACTAGCACAAGCTCTGGGACCGGACACACCATTTACGAGCATGGCCGGTTCGGAGATATTCTCTTTGGAAATGAGTAAAACTGAGGCTCTAACGCAGGCCATCAGGAAGTCTATAGGTATTAGAATTAAGGAGGAGTCTGAGATAATTGAAGGGGAGGTCGTCGAGGTGGTAGTGGAGCGTGCGGCGGGGGGCGGCGGCGCCAGGGTCGGCCGTCTCACCCTCAAGACCACCGACATGGAAACGAACTACGACATGGGAGCCAAAATGATTGATTCGCTGCTTAAAGAGAAGGTTCAAGCCGGAGATGTGATCACCATAGACAAGGCTACAGGCAAAATAAACAAGCTCGGCAGGAGTTTTGCCAGAGCCAGAGACTATGATGCTACCGGTCAGCAGGCCAGGTTCGTGCAGTGCCCCGAGGGCGAGCTGCAGAAGCGCAAGGAGGTGGTGCACACGGTGACGCTGCACGAGGTGGACGTCATCAACTCCAGGACCCACGGCTTCCTGGCACTGTTCTCCGGCGACACGGGCGAGATTAAATCTGAGATTCGGGAGCAGATTAACAGTAAGGTGGCTGAGTGGAGAGAGGAGGGGAAGGCAGAAATGATTCCGGGGGTGCTATTCATCGATGAGGCTCACATGCTGGACATTGAGTGCTTCTCGTTCCTCAATCGCGCGCTGGAATCGGAAACCGCTCCCATCGTGATCATGGCGACGAATCGAGGGATCACCCGGATACGGGGTACCACTTACCGCAGCCCGCACGGCATACCGCTGGACCTGCTCGACCGGATGATCATCGTGGCCACCTCGCCGTACTCCCAGAGTGAGCTCAAGGACATACTCAATATACGGTGA
Protein
MASIAAAQVQEVRSITRIERIGAHSHIRGLGLDDSLEPRQVSQGMVGQKMARKAAGVILQMIREGKIAGRAVLLAGQPGTGKTAIAMGLAQALGPDTPFTSMAGSEIFSLEMSKTEALTQAIRKSIGIRIKEESEIIEGEVVEVVVERAAGGGGARVGRLTLKTTDMETNYDMGAKMIDSLLKEKVQAGDVITIDKATGKINKLGRSFARARDYDATGQQARFVQCPEGELQKRKEVVHTVTLHEVDVINSRTHGFLALFSGDTGEIKSEIREQINSKVAEWREEGKAEMIPGVLFIDEAHMLDIECFSFLNRALESETAPIVIMATNRGITRIRGTTYRSPHGIPLDLLDRMIIVATSPYSQSELKDILNIR
Summary
Description
Proposed core component of the chromatin remodeling Ino80 complex which is involved in transcriptional regulation, DNA replication and probably DNA repair.
Proposed core component of the chromatin remodeling INO80 complex which is involved in transcriptional regulation, DNA replication and probably DNA repair.
Has single-stranded DNA-stimulated ATPase and ATP-dependent DNA helicase (5' to 3') activity suggesting a role in nuclear processes such as recombination and transcription.
Involved in the endoplasmic reticulum (ER)-associated degradation (ERAD) pathway where it negatively regulates expression of ER stress response genes.
Possesses single-stranded DNA-stimulated ATPase and ATP-dependent DNA helicase (5' to 3') activity; hexamerization is thought to be critical for ATP hydrolysis and adjacent subunits in the ring-like structure contribute to the ATPase activity.
Component of the NuA4 histone acetyltransferase complex which is involved in transcriptional activation of select genes principally by acetylation of nucleosomal histones H4 and H2A. This modification may both alter nucleosome - DNA interactions and promote interaction of the modified histones with other proteins which positively regulate transcription. This complex may be required for the activation of transcriptional programs associated with oncogene and proto-oncogene mediated growth induction, tumor suppressor mediated growth arrest and replicative senescence, apoptosis, and DNA repair. The NuA4 complex ATPase and helicase activities seem to be, at least in part, contributed by the association of RUVBL1 and RUVBL2 with EP400. NuA4 may also play a direct role in DNA repair when recruited to sites of DNA damage. Component of a SWR1-like complex that specifically mediates the removal of histone H2A.Z/H2AFZ from the nucleosome (By similarity).
Plays an essential role in oncogenic transformation by MYC and also modulates transcriptional activation by the LEF1/TCF1-CTNNB1 complex. May also inhibit the transcriptional activity of ATF2 (By similarity).
Proposed core component of the chromatin remodeling INO80 complex which is involved in transcriptional regulation, DNA replication and probably DNA repair.
Has single-stranded DNA-stimulated ATPase and ATP-dependent DNA helicase (5' to 3') activity suggesting a role in nuclear processes such as recombination and transcription.
Involved in the endoplasmic reticulum (ER)-associated degradation (ERAD) pathway where it negatively regulates expression of ER stress response genes.
Possesses single-stranded DNA-stimulated ATPase and ATP-dependent DNA helicase (5' to 3') activity; hexamerization is thought to be critical for ATP hydrolysis and adjacent subunits in the ring-like structure contribute to the ATPase activity.
Component of the NuA4 histone acetyltransferase complex which is involved in transcriptional activation of select genes principally by acetylation of nucleosomal histones H4 and H2A. This modification may both alter nucleosome - DNA interactions and promote interaction of the modified histones with other proteins which positively regulate transcription. This complex may be required for the activation of transcriptional programs associated with oncogene and proto-oncogene mediated growth induction, tumor suppressor mediated growth arrest and replicative senescence, apoptosis, and DNA repair. The NuA4 complex ATPase and helicase activities seem to be, at least in part, contributed by the association of RUVBL1 and RUVBL2 with EP400. NuA4 may also play a direct role in DNA repair when recruited to sites of DNA damage. Component of a SWR1-like complex that specifically mediates the removal of histone H2A.Z/H2AFZ from the nucleosome (By similarity).
Plays an essential role in oncogenic transformation by MYC and also modulates transcriptional activation by the LEF1/TCF1-CTNNB1 complex. May also inhibit the transcriptional activity of ATF2 (By similarity).
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Subunit
Forms homohexameric rings (Probable). Can form a dodecamer with ruvbl2 made of two stacked hexameric rings. Is a component of the RNA polymerase II holoenzyme complex. Component of the chromatin-remodeling Ino80 complex. Component of some MLL1/MLL complex (By similarity).
Forms homohexameric rings (Probable). Can form a dodecamer with RUVBL1 made of two stacked hexameric rings; however, even though RUVBL1 and RUVBL2 are present in equimolar ratio, the oligomeric status of each hexamer is not known. Oligomerization may regulate binding to nucleic acids and conversely, binding to nucleic acids may affect the dodecameric assembly. Interacts with the transcriptional activation domain of MYC. Interacts With ATF2. Component of the RNA polymerase II holoenzyme complex. May also act to bridge the LEF1/TCF1-CTNNB1 complex and TBP. Component of the NuA4 histone acetyltransferase complex which contains the catalytic subunit KAT5/TIP60 and the subunits EP400, TRRAP/PAF400, BRD8/SMAP, EPC1, DMAP1/DNMAP1, RUVBL1/TIP49, RUVBL2, ING3, actin, ACTL6A/BAF53A, MORF4L1/MRG15, MORF4L2/MRGX, MRGBP, YEATS4/GAS41, VPS72/YL1 and MEAF6. The NuA4 complex interacts with MYC and the adenovirus E1A protein. RUVBL2 interacts with EP400. Component of a NuA4-related complex which contains EP400, TRRAP/PAF400, SRCAP, BRD8/SMAP, EPC1, DMAP1/DNMAP1, RUVBL1/TIP49, RUVBL2, actin, ACTL6A/BAF53A, VPS72 and YEATS4/GAS41. Interacts with NPAT. Component of the chromatin-remodeling INO80 complex; specifically part of a complex module associated with the helicase ATP-binding and the helicase C-terminal domain of INO80. Component of some MLL1/MLL complex, at least composed of the core components KMT2A/MLL1, ASH2L, HCFC1/HCF1, WDR5 and RBBP5, as well as the facultative components BAP18, CHD8, E2F6, HSP70, INO80C, KANSL1, LAS1L, MAX, MCRS1, MGA, MYST1/MOF, PELP1, PHF20, PRP31, RING2, RUVB1/TIP49A, RUVB2/TIP49B, SENP3, TAF1, TAF4, TAF6, TAF7, TAF9 and TEX10. Interacts with IGHMBP2. Interacts with TELO2. Interacts with HINT1. Component of a SWR1-like complex. Component of the R2TP complex composed at least of PIHD1, RUVBL1, RUVBL2 and RPAP3. Interacts with ITFG1. Interacts with ZMYND10. Interacts with WAC; WAC positively regulates MTOR activity by promoting the assembly of the TTT complex composed of TELO2, TTI1 and TTI2 and the RUVBL complex composed of RUVBL1 and RUVBL2 into the TTT-RUVBL complex which leads to the dimerization of the mTORC1 complex and its subsequent activation. Forms a complex with APPL1 and APPL2 (By similarity).
Forms homohexameric rings (Probable). Can form a dodecamer with RUVBL1 made of two stacked hexameric rings; however, even though RUVBL1 and RUVBL2 are present in equimolar ratio, the oligomeric status of each hexamer is not known. Oligomerization may regulate binding to nucleic acids and conversely, binding to nucleic acids may affect the dodecameric assembly. Interacts with the transcriptional activation domain of MYC. Interacts With ATF2. Component of the RNA polymerase II holoenzyme complex. May also act to bridge the LEF1/TCF1-CTNNB1 complex and TBP. Component of the NuA4 histone acetyltransferase complex which contains the catalytic subunit KAT5/TIP60 and the subunits EP400, TRRAP/PAF400, BRD8/SMAP, EPC1, DMAP1/DNMAP1, RUVBL1/TIP49, RUVBL2, ING3, actin, ACTL6A/BAF53A, MORF4L1/MRG15, MORF4L2/MRGX, MRGBP, YEATS4/GAS41, VPS72/YL1 and MEAF6. The NuA4 complex interacts with MYC and the adenovirus E1A protein. RUVBL2 interacts with EP400. Component of a NuA4-related complex which contains EP400, TRRAP/PAF400, SRCAP, BRD8/SMAP, EPC1, DMAP1/DNMAP1, RUVBL1/TIP49, RUVBL2, actin, ACTL6A/BAF53A, VPS72 and YEATS4/GAS41. Interacts with NPAT. Component of the chromatin-remodeling INO80 complex; specifically part of a complex module associated with the helicase ATP-binding and the helicase C-terminal domain of INO80. Component of some MLL1/MLL complex, at least composed of the core components KMT2A/MLL1, ASH2L, HCFC1/HCF1, WDR5 and RBBP5, as well as the facultative components BAP18, CHD8, E2F6, HSP70, INO80C, KANSL1, LAS1L, MAX, MCRS1, MGA, MYST1/MOF, PELP1, PHF20, PRP31, RING2, RUVB1/TIP49A, RUVB2/TIP49B, SENP3, TAF1, TAF4, TAF6, TAF7, TAF9 and TEX10. Interacts with IGHMBP2. Interacts with TELO2. Interacts with HINT1. Component of a SWR1-like complex. Component of the R2TP complex composed at least of PIHD1, RUVBL1, RUVBL2 and RPAP3. Interacts with ITFG1. Interacts with ZMYND10. Interacts with WAC; WAC positively regulates MTOR activity by promoting the assembly of the TTT complex composed of TELO2, TTI1 and TTI2 and the RUVBL complex composed of RUVBL1 and RUVBL2 into the TTT-RUVBL complex which leads to the dimerization of the mTORC1 complex and its subsequent activation. Forms a complex with APPL1 and APPL2 (By similarity).
Similarity
Belongs to the RuvB family.
Keywords
ATP-binding
DNA damage
DNA recombination
DNA repair
Helicase
Hydrolase
Nucleotide-binding
Nucleus
Transcription
Transcription regulation
Acetylation
Activator
Chromatin regulator
Complete proteome
Isopeptide bond
Phosphoprotein
Reference proteome
Ubl conjugation
Feature
chain RuvB-like helicase
Uniprot
A0A2A4K490
A0A0N0PC54
A0A2W1BK50
A0A1B6GNH1
A0A1B6IPC2
A0A2A3ELN2
+ More
A0A088ARN2 A0A0L7RDH9 A0A0M8ZWR1 A0A1B6D3M9 A0A154PA15 A0A0C9QK39 A0A310SDU0 E2B6H7 A0A2J7RI53 A0A151IGB9 E9IXY8 A0A067QZM3 A0A195DCE3 A0A0J7K820 E0VPV9 A0A158NEA8 A0A2J7RI55 A0A151WFD9 A0A0J7KGE9 F4WM57 A0A232FD79 K7IZH3 A0A026WXH9 E9GRY1 A0A195FUP0 A0A2R5LM78 G1KMP5 Q0IH85 A0A0P5UL26 A0A0B8RWC7 G3MNC5 A0A0P5DJ95 A0A0P5UNC6 A0A0P5NVU9 A0A0P5ULG6 Q801R1 A0A1W7RDN6 T1DL73 A0A0F7Z5R1 A0A0P5AGE5 Q9DE27 A0A0P6H0Y4 A0A0P5YYQ6 A0A0N8DWN2 Q28GB7 A0A224YYM6 A0A131YT57 L7M846 A0A0P5HQ08 A0A0F7YZJ1 A0A2H6NAJ8 A0A1S3JEQ9 A0A131XH09 A0A0P6F024 A0A0P4ZBA1 C3Y6A4 A0A2K5QG80 A0A2D4NPV6 A0A0P5JVV9 A0A0P5BH73 A0A2K5EYP0 A0A0P5QCE6 A0A2K5EYU9 F6UB78 A0A2K6U4D8 F7GQG2 A0A151NKW1 A0A286XN46 A0A061I456 A0A287D5H8 A0A1U7RDA2 G3V8T5 Q3TXT7 F6X277 A0A2Y9F1Z3 A0A1S3JER7 Q9WTM5 V9KTC5 A0A1S3G4W2 Q4QQS4 A0A2Y9F0F3 A0A1A7YLD1 I3MM17 L5LT14 A0A3Q2D7M0 A0A384BKV9 A0A3Q0DWP0 A0A3Q7TNI3 A0A0P5S644 A0A146WVK1 A0A3Q2U7I7 G1QY99 A0A146WSZ4 H0W8I2
A0A088ARN2 A0A0L7RDH9 A0A0M8ZWR1 A0A1B6D3M9 A0A154PA15 A0A0C9QK39 A0A310SDU0 E2B6H7 A0A2J7RI53 A0A151IGB9 E9IXY8 A0A067QZM3 A0A195DCE3 A0A0J7K820 E0VPV9 A0A158NEA8 A0A2J7RI55 A0A151WFD9 A0A0J7KGE9 F4WM57 A0A232FD79 K7IZH3 A0A026WXH9 E9GRY1 A0A195FUP0 A0A2R5LM78 G1KMP5 Q0IH85 A0A0P5UL26 A0A0B8RWC7 G3MNC5 A0A0P5DJ95 A0A0P5UNC6 A0A0P5NVU9 A0A0P5ULG6 Q801R1 A0A1W7RDN6 T1DL73 A0A0F7Z5R1 A0A0P5AGE5 Q9DE27 A0A0P6H0Y4 A0A0P5YYQ6 A0A0N8DWN2 Q28GB7 A0A224YYM6 A0A131YT57 L7M846 A0A0P5HQ08 A0A0F7YZJ1 A0A2H6NAJ8 A0A1S3JEQ9 A0A131XH09 A0A0P6F024 A0A0P4ZBA1 C3Y6A4 A0A2K5QG80 A0A2D4NPV6 A0A0P5JVV9 A0A0P5BH73 A0A2K5EYP0 A0A0P5QCE6 A0A2K5EYU9 F6UB78 A0A2K6U4D8 F7GQG2 A0A151NKW1 A0A286XN46 A0A061I456 A0A287D5H8 A0A1U7RDA2 G3V8T5 Q3TXT7 F6X277 A0A2Y9F1Z3 A0A1S3JER7 Q9WTM5 V9KTC5 A0A1S3G4W2 Q4QQS4 A0A2Y9F0F3 A0A1A7YLD1 I3MM17 L5LT14 A0A3Q2D7M0 A0A384BKV9 A0A3Q0DWP0 A0A3Q7TNI3 A0A0P5S644 A0A146WVK1 A0A3Q2U7I7 G1QY99 A0A146WSZ4 H0W8I2
EC Number
3.6.4.12
Pubmed
26354079
28756777
20798317
21282665
24845553
20566863
+ More
21347285 21719571 28648823 20075255 24508170 30249741 21292972 27762356 25476704 22216098 26358130 23758969 10842076 20431018 28797301 26830274 25576852 28049606 18563158 17495919 25243066 22293439 21993624 23929341 15057822 15632090 10349636 11042159 11076861 11217851 12466851 12040188 15489334 16141073 21183079 19892987 10220587 24402279 24813606 15592404
21347285 21719571 28648823 20075255 24508170 30249741 21292972 27762356 25476704 22216098 26358130 23758969 10842076 20431018 28797301 26830274 25576852 28049606 18563158 17495919 25243066 22293439 21993624 23929341 15057822 15632090 10349636 11042159 11076861 11217851 12466851 12040188 15489334 16141073 21183079 19892987 10220587 24402279 24813606 15592404
EMBL
NWSH01000147
PCG79067.1
KQ460772
KPJ12452.1
KZ149995
PZC75452.1
+ More
GECZ01005804 JAS63965.1 GECU01018924 JAS88782.1 KZ288222 PBC32106.1 KQ414614 KOC68864.1 KQ435814 KOX72680.1 GEDC01017004 GEDC01007112 JAS20294.1 JAS30186.1 KQ434856 KZC08735.1 GBYB01000972 JAG70739.1 KQ760869 OAD58769.1 GL445973 EFN88721.1 NEVH01003505 PNF40512.1 KQ977726 KYN00309.1 GL766836 EFZ14571.1 KK852809 KDR16004.1 KQ980989 KYN10580.1 LBMM01012069 KMQ86429.1 DS235379 EEB15415.1 ADTU01001328 PNF40514.1 KQ983219 KYQ46550.1 LBMM01007939 KMQ89281.1 GL888217 EGI64701.1 NNAY01000430 OXU28453.1 KK107069 QOIP01000002 EZA60732.1 RLU25975.1 GL732561 EFX77610.1 KQ981264 KYN44007.1 GGLE01006496 MBY10622.1 BC123265 CM004478 AAI23266.1 OCT73146.1 GDIP01111401 JAL92313.1 GBSH01002404 JAG66622.1 JO843376 AEO34993.1 GDIP01161390 JAJ62012.1 GDIP01112601 JAL91113.1 GDIQ01144085 JAL07641.1 GDIP01111400 JAL92314.1 BC047966 AAH47966.1 GDAY02002171 JAV49261.1 GAAZ01002212 JAA95731.1 GBEX01003007 JAI11553.1 GDIP01202286 JAJ21116.1 AF218071 GDIQ01026112 JAN68625.1 GDIP01051412 JAM52303.1 GDIP01073002 GDIQ01084612 JAM30713.1 JAN10125.1 AAMC01090096 CR761457 CAJ83917.1 GFPF01011560 MAA22706.1 GEDV01006887 JAP81670.1 GACK01004834 JAA60200.1 GDIQ01224698 JAK27027.1 GBEW01000909 JAI09456.1 IACI01072922 LAA28282.1 GEFH01003580 JAP65001.1 GDIQ01063304 JAN31433.1 GDIP01216533 JAJ06869.1 GG666487 EEN64501.1 IACN01014288 LAB47740.1 GDIQ01194452 JAK57273.1 GDIP01189983 JAJ33419.1 GDIQ01116298 JAL35428.1 GAMT01008890 GAMS01009298 GAMR01004745 GAMQ01005695 GAMP01009703 JAB02971.1 JAB13838.1 JAB29187.1 JAB36156.1 JAB43052.1 AKHW03002822 KYO37125.1 AAKN02046418 AAKN02046419 KE680915 ERE71143.1 AGTP01089690 AC128792 CH473979 EDM07354.1 BC115809 BC116693 AK144156 AK159112 CH466603 AAI15810.1 BAE34828.1 EDL22854.1 AB013912 JW868924 AFP01442.1 BC098042 AAH98042.1 HADW01019644 HADX01009200 SBP31432.1 KB109009 ELK28598.1 GDIQ01108990 JAL42736.1 GCES01053286 JAR33037.1 AADN05000747 ADFV01051887 ADFV01051888 ADFV01051889 ADFV01051890 ADFV01051891 GCES01053283 JAR33040.1
GECZ01005804 JAS63965.1 GECU01018924 JAS88782.1 KZ288222 PBC32106.1 KQ414614 KOC68864.1 KQ435814 KOX72680.1 GEDC01017004 GEDC01007112 JAS20294.1 JAS30186.1 KQ434856 KZC08735.1 GBYB01000972 JAG70739.1 KQ760869 OAD58769.1 GL445973 EFN88721.1 NEVH01003505 PNF40512.1 KQ977726 KYN00309.1 GL766836 EFZ14571.1 KK852809 KDR16004.1 KQ980989 KYN10580.1 LBMM01012069 KMQ86429.1 DS235379 EEB15415.1 ADTU01001328 PNF40514.1 KQ983219 KYQ46550.1 LBMM01007939 KMQ89281.1 GL888217 EGI64701.1 NNAY01000430 OXU28453.1 KK107069 QOIP01000002 EZA60732.1 RLU25975.1 GL732561 EFX77610.1 KQ981264 KYN44007.1 GGLE01006496 MBY10622.1 BC123265 CM004478 AAI23266.1 OCT73146.1 GDIP01111401 JAL92313.1 GBSH01002404 JAG66622.1 JO843376 AEO34993.1 GDIP01161390 JAJ62012.1 GDIP01112601 JAL91113.1 GDIQ01144085 JAL07641.1 GDIP01111400 JAL92314.1 BC047966 AAH47966.1 GDAY02002171 JAV49261.1 GAAZ01002212 JAA95731.1 GBEX01003007 JAI11553.1 GDIP01202286 JAJ21116.1 AF218071 GDIQ01026112 JAN68625.1 GDIP01051412 JAM52303.1 GDIP01073002 GDIQ01084612 JAM30713.1 JAN10125.1 AAMC01090096 CR761457 CAJ83917.1 GFPF01011560 MAA22706.1 GEDV01006887 JAP81670.1 GACK01004834 JAA60200.1 GDIQ01224698 JAK27027.1 GBEW01000909 JAI09456.1 IACI01072922 LAA28282.1 GEFH01003580 JAP65001.1 GDIQ01063304 JAN31433.1 GDIP01216533 JAJ06869.1 GG666487 EEN64501.1 IACN01014288 LAB47740.1 GDIQ01194452 JAK57273.1 GDIP01189983 JAJ33419.1 GDIQ01116298 JAL35428.1 GAMT01008890 GAMS01009298 GAMR01004745 GAMQ01005695 GAMP01009703 JAB02971.1 JAB13838.1 JAB29187.1 JAB36156.1 JAB43052.1 AKHW03002822 KYO37125.1 AAKN02046418 AAKN02046419 KE680915 ERE71143.1 AGTP01089690 AC128792 CH473979 EDM07354.1 BC115809 BC116693 AK144156 AK159112 CH466603 AAI15810.1 BAE34828.1 EDL22854.1 AB013912 JW868924 AFP01442.1 BC098042 AAH98042.1 HADW01019644 HADX01009200 SBP31432.1 KB109009 ELK28598.1 GDIQ01108990 JAL42736.1 GCES01053286 JAR33037.1 AADN05000747 ADFV01051887 ADFV01051888 ADFV01051889 ADFV01051890 ADFV01051891 GCES01053283 JAR33040.1
Proteomes
UP000218220
UP000053240
UP000242457
UP000005203
UP000053825
UP000053105
+ More
UP000076502 UP000008237 UP000235965 UP000078542 UP000027135 UP000078492 UP000036403 UP000009046 UP000005205 UP000075809 UP000007755 UP000215335 UP000002358 UP000053097 UP000279307 UP000000305 UP000078541 UP000001646 UP000186698 UP000008143 UP000085678 UP000001554 UP000233040 UP000233020 UP000002280 UP000233220 UP000050525 UP000005447 UP000030759 UP000005215 UP000189706 UP000002494 UP000002281 UP000248484 UP000000589 UP000081671 UP000265020 UP000261680 UP000291021 UP000189704 UP000286642 UP000000539 UP000001073
UP000076502 UP000008237 UP000235965 UP000078542 UP000027135 UP000078492 UP000036403 UP000009046 UP000005205 UP000075809 UP000007755 UP000215335 UP000002358 UP000053097 UP000279307 UP000000305 UP000078541 UP000001646 UP000186698 UP000008143 UP000085678 UP000001554 UP000233040 UP000233020 UP000002280 UP000233220 UP000050525 UP000005447 UP000030759 UP000005215 UP000189706 UP000002494 UP000002281 UP000248484 UP000000589 UP000081671 UP000265020 UP000261680 UP000291021 UP000189704 UP000286642 UP000000539 UP000001073
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A2A4K490
A0A0N0PC54
A0A2W1BK50
A0A1B6GNH1
A0A1B6IPC2
A0A2A3ELN2
+ More
A0A088ARN2 A0A0L7RDH9 A0A0M8ZWR1 A0A1B6D3M9 A0A154PA15 A0A0C9QK39 A0A310SDU0 E2B6H7 A0A2J7RI53 A0A151IGB9 E9IXY8 A0A067QZM3 A0A195DCE3 A0A0J7K820 E0VPV9 A0A158NEA8 A0A2J7RI55 A0A151WFD9 A0A0J7KGE9 F4WM57 A0A232FD79 K7IZH3 A0A026WXH9 E9GRY1 A0A195FUP0 A0A2R5LM78 G1KMP5 Q0IH85 A0A0P5UL26 A0A0B8RWC7 G3MNC5 A0A0P5DJ95 A0A0P5UNC6 A0A0P5NVU9 A0A0P5ULG6 Q801R1 A0A1W7RDN6 T1DL73 A0A0F7Z5R1 A0A0P5AGE5 Q9DE27 A0A0P6H0Y4 A0A0P5YYQ6 A0A0N8DWN2 Q28GB7 A0A224YYM6 A0A131YT57 L7M846 A0A0P5HQ08 A0A0F7YZJ1 A0A2H6NAJ8 A0A1S3JEQ9 A0A131XH09 A0A0P6F024 A0A0P4ZBA1 C3Y6A4 A0A2K5QG80 A0A2D4NPV6 A0A0P5JVV9 A0A0P5BH73 A0A2K5EYP0 A0A0P5QCE6 A0A2K5EYU9 F6UB78 A0A2K6U4D8 F7GQG2 A0A151NKW1 A0A286XN46 A0A061I456 A0A287D5H8 A0A1U7RDA2 G3V8T5 Q3TXT7 F6X277 A0A2Y9F1Z3 A0A1S3JER7 Q9WTM5 V9KTC5 A0A1S3G4W2 Q4QQS4 A0A2Y9F0F3 A0A1A7YLD1 I3MM17 L5LT14 A0A3Q2D7M0 A0A384BKV9 A0A3Q0DWP0 A0A3Q7TNI3 A0A0P5S644 A0A146WVK1 A0A3Q2U7I7 G1QY99 A0A146WSZ4 H0W8I2
A0A088ARN2 A0A0L7RDH9 A0A0M8ZWR1 A0A1B6D3M9 A0A154PA15 A0A0C9QK39 A0A310SDU0 E2B6H7 A0A2J7RI53 A0A151IGB9 E9IXY8 A0A067QZM3 A0A195DCE3 A0A0J7K820 E0VPV9 A0A158NEA8 A0A2J7RI55 A0A151WFD9 A0A0J7KGE9 F4WM57 A0A232FD79 K7IZH3 A0A026WXH9 E9GRY1 A0A195FUP0 A0A2R5LM78 G1KMP5 Q0IH85 A0A0P5UL26 A0A0B8RWC7 G3MNC5 A0A0P5DJ95 A0A0P5UNC6 A0A0P5NVU9 A0A0P5ULG6 Q801R1 A0A1W7RDN6 T1DL73 A0A0F7Z5R1 A0A0P5AGE5 Q9DE27 A0A0P6H0Y4 A0A0P5YYQ6 A0A0N8DWN2 Q28GB7 A0A224YYM6 A0A131YT57 L7M846 A0A0P5HQ08 A0A0F7YZJ1 A0A2H6NAJ8 A0A1S3JEQ9 A0A131XH09 A0A0P6F024 A0A0P4ZBA1 C3Y6A4 A0A2K5QG80 A0A2D4NPV6 A0A0P5JVV9 A0A0P5BH73 A0A2K5EYP0 A0A0P5QCE6 A0A2K5EYU9 F6UB78 A0A2K6U4D8 F7GQG2 A0A151NKW1 A0A286XN46 A0A061I456 A0A287D5H8 A0A1U7RDA2 G3V8T5 Q3TXT7 F6X277 A0A2Y9F1Z3 A0A1S3JER7 Q9WTM5 V9KTC5 A0A1S3G4W2 Q4QQS4 A0A2Y9F0F3 A0A1A7YLD1 I3MM17 L5LT14 A0A3Q2D7M0 A0A384BKV9 A0A3Q0DWP0 A0A3Q7TNI3 A0A0P5S644 A0A146WVK1 A0A3Q2U7I7 G1QY99 A0A146WSZ4 H0W8I2
PDB
6QI9
E-value=1.29105e-143,
Score=1306
Ontologies
GO
GO:0043141
GO:0097255
GO:0035267
GO:0006281
GO:0005524
GO:0006325
GO:0031011
GO:0000812
GO:0006357
GO:0006338
GO:0004003
GO:0016573
GO:0000492
GO:0005829
GO:0008013
GO:0071733
GO:0000980
GO:0043967
GO:0071169
GO:0071339
GO:0090090
GO:0005719
GO:0071899
GO:0000979
GO:0031490
GO:0017025
GO:0042802
GO:0035066
GO:0034644
GO:0003714
GO:0005813
GO:0006310
GO:0071392
GO:0043968
GO:0044458
GO:0003351
GO:0048565
GO:0045892
GO:0007507
GO:0060420
GO:0070286
GO:0051117
GO:0043531
GO:0001094
GO:0042803
GO:1990904
GO:0045944
GO:0005737
GO:0005654
GO:0005634
GO:0003678
GO:0004386
GO:0006412
GO:0016020
GO:0022857
GO:0004559
GO:0006013
GO:0015923
GO:0006260
PANTHER
Topology
Subcellular location
Nucleus
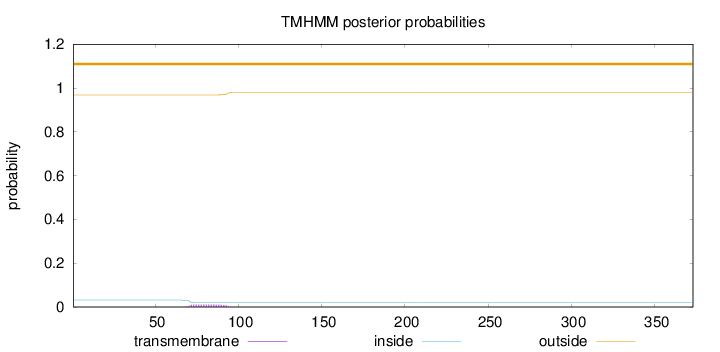
Length:
373
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.25202
Exp number, first 60 AAs:
0.00048
Total prob of N-in:
0.03175
outside
1 - 373
Population Genetic Test Statistics
Pi
274.824049
Theta
187.444091
Tajima's D
1.570267
CLR
0.236031
CSRT
0.808559572021399
Interpretation
Uncertain
