Gene
KWMTBOMO06230
Pre Gene Modal
BGIBMGA001641
Annotation
Leucyl-cystinyl_aminopeptidase_[Papilio_xuthus]
Full name
Aminopeptidase
Location in the cell
Mitochondrial Reliability : 1.287 PlasmaMembrane Reliability : 1.05
Sequence
CDS
ATGGAAGGTATTCCGATGGAGCCAATAGGCAAATACAAAGCAGACAGGAACGGTTCAGCGAAAGCAGCGGCCTGGCGTAATATGGTCGATCAAGATCTGGATGACGTGGCATTTCTAGCTGACACAGATAAGGCGCGTGAAGCCAGAGAGCTCCGCGACGCTCCCGTGGCCGTCTGCTCACAGCGCCGAGCGGTTTGCCTCACTGTTCTGGCCTTCGCTACGATCTTCGCCACGTCCTTGCTGGTCGTGTACGCCAGTCCACAGCCGGAATGTCCGTGTGCTGAGGAAACGACTTTAATAGTTGGACAGCCACCGACGGACGCGGATAACGTGGCTTCATCAGCTAACAAAGAACGCATCGCTAGCAATGGCGCGGTGTTCCCGTGGCGCGGTGCGAGGTTGCCAACGTTCGTGATACCAAAACACTACAGTCTGTGGCTGCATCCGAATCTGACCACAGGCGAATTGAGAGGTGAAGTTTCGATTGACCTGAAGGTGGACAGAGACACCACGTTTGTGGTGCTCAACGTGAGAGACATGAACGTGACCGAACGGGCGCTCTTCAAGTCGGGAGGATCTCTGGGACCGAAAATCTCTAGGGTCTTAGATTACCCGCAAGCCGATCAGACTTACATTGAATTTAAGGAGAAAATACGAAGAAAATATAACTACACATTGAGTTTGCGTTTCATCACGAGATTGGAGAGGAGTGACAAGCAGAGAGGCTTCTTCTTGACCGGAAACCAAAGACATCGCTGCGCTGTCTCCAGATTTTGGTTGACCCACGCACGATCCACTTTCCCTTGTTTCGATGAGCCCAATTTAAGGGCCAGCTTCAAGCTGACCATAGTAAGAGACAGGTTCCACGTGTCTCTTACGAACATGCCCATAGTTGCGACAGAAGAAGCGGGATTCTATCTTGGACACAGACTCCTCCAGGACGAATTCGCCACCTCGCCCCCCATGCCCCCCCACATGGTGGCGGTGGCGGTCTGCCGTCTCCAAAGAGCGTCAGCGCCGACACCGGAGGCTAACACAACCGACGCCACCGAAGCGGAGACCGACGGAGATACCAGCACAGCACCAGAGATCAGCCTTTACACTGACCACCCGAGCATATTACAGGAGAGCGGACCGCTTCTCGAATGGTTGCAGAAAACCATCCAGCAGTTCAGTTACGAGTTGAACACGTCGTATCCTCTACCGAAATTCGATGTCGTGGTGGTTGACAGCGCCAACCACTATTCAGAAGGTTGGGGCCTCATTACTCTCGCGCCAGCCACTTTATCTGACACCAAAACTATAGCAAGACTTTTGGCACAGCAGTGGTTCGGAGGCCTAGTCTCTCCAAGATGGTGGGCGTCGCAGTGGCTGATGGAAGCCCTAACATCGCTAATAGCTGAAAAAGCACCGCCCTTCAAAAATTCCGCCCTGAAGCAAGAAGAAGCTTTGCTGCTCGACCATGTTCTGCCAGCACTGAGGTTAGACTCTGGAAGTACAGTTCGGGCGGTCGCGGCATTGAGACTCGAACGCGCCGACATCGAGTCGGCTACGGATGAACTCTCTCTGCACAAGGGTGCTGCCATAGTTAGTATGGCAATCGAAGCAGCCGGTGAGGAAACTGGCCGAGCAGCACTCGCGCGTCTGTTGCGCGATCACCGCTATGCAAGCGCCGACGCCAGAGATTTATGGCGGGCGTTGCAACGTGACAACCCTGACGATGCATCGGTGCAGGCCCACGCGTGGGACGGCTGGTGCGAGAAGCCAGGGTATCCGTTGCTGAGCGCGACGACAGTCGGAGATGACGTCATTCTCAAACAAGAAAGGTTCCTGATGACGGCCGAACCACCACCCCCAGAGCCTCCAATGCTGAACGACCTCCTCACTCAGGACTTGAGAGCCGATCTCGAAGCATTGTTCTTTGATCCAGAAAACTACACGGACGCTACCGAAGAGGACTACAATGCCACCACCACGACCACCCCACTCCCAACCACCACCACCAAGAAGCCGTCGCCGAAGACGACGAAGCTGCCGCCACCGAAATGGATTATCCCGATTACCTTCACCGTTGGCCCCCTGGAAGCAGACCCGGAGGAAATCCACACAATGCACAACGGAACTTGGTACGACATTGGAAACGAGACAAAGCCGCTTCAGACATCGCGATGGACGGAGAACACGACGCATCTGGTCTGGATGAACGATACCGAAATGGTCATACCGGATCTAGGTAAACACAAATGGATCAGATACAACGTCGGAGCTCGTGGACTGTACCGGGTCGCGCCACAGGACCGAGCAGGGGAGGAGGCTTCCGATGCTGCTCGCGTTTACGATGGCGCTTCGGCCGCTGAAAGAGCCTTGATTTTAGATGATGCCTTCGTCCTGAGCCGAGCCGGGAGACTGCCGGCCAGTCGAGCTATAGTCGTAGCTGCCCGGATCCGGGGCGAACAGCACTGGGCTCCGTGGCGAGTGGTCCTCTCGCACATGTCGTGGTGGCGCGAGTTGCTCAGGGAAGGGGCAGCCGCTCCCGCCCTAGCGCGTCTGCTGGCCACGTTGCACCCACCGATCGCGCTGCGTCACCAACGCGACGCCGACTCCGACGATCACTTGTGGCTGCGCGGAGCCCTCCTCGCGTCCGGAGTGGAGTGGGGCAATCAAGGGATCACGAACGAAGCCGTACAGTTGTTCGACCTCTGGATGGAGAAAAATCACACGATACCGGAGATATACCAGGAGGCGGCGTTCACTGCGGGCGTTCGGACACACGGGCGGGTCGCGTGGCGCGCGTGCTGGCGAGCACTCGTGGACTCGTATTCGGCGCCGCGCCCTACATACAGCCACAGAGCCCTGCTGGCGGCGCTCGCGTCGCCCGAAGACGACTGGCTGTTCTACAGGTTCGCGTTCACCGTCCTCTCGACGGAGGCTCAGCGTGGCCGCGACTGGACTGAGTGGATCACCGCACTCTACACGAGCACATGCAGATGGCGCGGCGCTGCTGGCATTTGGAGGATCCTAAGACCTGAAACCTCGCTACCAGCGTCGGCGCTCCAAGCAGCCGCCCGCTGTCTCCACCAGCCCTACGACTATTATCGGTTTAAGGAGTTGTTCGGTGAGCTTCGAGATTCGCAATCGTGGCTGGACACGATAGCCCTGAACGCTGCCTGGGTGTCCAAAGCGGATTCGGACCTGGCCCGATATTTCGCGACCCTACACGACCCACAGAGCCATTAG
Protein
MEGIPMEPIGKYKADRNGSAKAAAWRNMVDQDLDDVAFLADTDKAREARELRDAPVAVCSQRRAVCLTVLAFATIFATSLLVVYASPQPECPCAEETTLIVGQPPTDADNVASSANKERIASNGAVFPWRGARLPTFVIPKHYSLWLHPNLTTGELRGEVSIDLKVDRDTTFVVLNVRDMNVTERALFKSGGSLGPKISRVLDYPQADQTYIEFKEKIRRKYNYTLSLRFITRLERSDKQRGFFLTGNQRHRCAVSRFWLTHARSTFPCFDEPNLRASFKLTIVRDRFHVSLTNMPIVATEEAGFYLGHRLLQDEFATSPPMPPHMVAVAVCRLQRASAPTPEANTTDATEAETDGDTSTAPEISLYTDHPSILQESGPLLEWLQKTIQQFSYELNTSYPLPKFDVVVVDSANHYSEGWGLITLAPATLSDTKTIARLLAQQWFGGLVSPRWWASQWLMEALTSLIAEKAPPFKNSALKQEEALLLDHVLPALRLDSGSTVRAVAALRLERADIESATDELSLHKGAAIVSMAIEAAGEETGRAALARLLRDHRYASADARDLWRALQRDNPDDASVQAHAWDGWCEKPGYPLLSATTVGDDVILKQERFLMTAEPPPPEPPMLNDLLTQDLRADLEALFFDPENYTDATEEDYNATTTTTPLPTTTTKKPSPKTTKLPPPKWIIPITFTVGPLEADPEEIHTMHNGTWYDIGNETKPLQTSRWTENTTHLVWMNDTEMVIPDLGKHKWIRYNVGARGLYRVAPQDRAGEEASDAARVYDGASAAERALILDDAFVLSRAGRLPASRAIVVAARIRGEQHWAPWRVVLSHMSWWRELLREGAAAPALARLLATLHPPIALRHQRDADSDDHLWLRGALLASGVEWGNQGITNEAVQLFDLWMEKNHTIPEIYQEAAFTAGVRTHGRVAWRACWRALVDSYSAPRPTYSHRALLAALASPEDDWLFYRFAFTVLSTEAQRGRDWTEWITALYTSTCRWRGAAGIWRILRPETSLPASALQAAARCLHQPYDYYRFKELFGELRDSQSWLDTIALNAAWVSKADSDLARYFATLHDPQSH
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M1 family.
Uniprot
EC Number
3.4.11.-
EMBL
Proteomes
Interpro
Gene 3D
CDD
ProteinModelPortal
PDB
5C97
E-value=1.95121e-47,
Score=481
Ontologies
GO
PANTHER
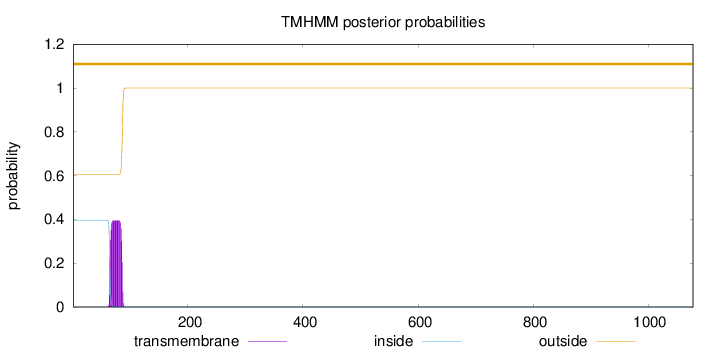
Topology
Length:
1078
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.56678999999996
Exp number, first 60 AAs:
0.00107
Total prob of N-in:
0.39496
outside
1 - 1078
Population Genetic Test Statistics
Pi
209.108601
Theta
170.257279
Tajima's D
0.906875
CLR
0.112148
CSRT
0.631918404079796
Interpretation
Uncertain
