Gene
KWMTBOMO06225
Pre Gene Modal
BGIBMGA001799
Annotation
PREDICTED:_ATP-dependent_RNA_helicase_abstrakt_[Amyelois_transitella]
Full name
ATP-dependent RNA helicase abstrakt
Location in the cell
Cytoplasmic Reliability : 1.774 Mitochondrial Reliability : 1.815
Sequence
CDS
ATGTCAGATGTAAAAGATGATCCAGCCCCTCCGCCGCCGATTAAACGGTACAGGCGCGAGGAAAAATCAGATTCTGACGAAGACGTGGACAACTATGAACCTTACGTTCCCATCAAAGAACGTAAGAAACAGCAGCTTTTGAAGCTAGGAAGGCTCGGGCTGCTCACTGCGGAAGCTGCGGCGGACACTAAAAGCTCTAGCGAAAACGACCATGACGATGAAACATCTCAAGAGGAATGGGGCAGACGTTACAATGTTTCATTGCTGGACCAGCACAGTGAGTTGAAGAGACTGGCCGAGGCCCGAGCTCTCTCCGCAGCCGAAAAACAGGCAAGAGAAGAGGAACATATACTCGAGAGTGTAGCGCAGAGCAAAGCCCTGATGGGAGTGGCGGAGTTGGCTAAAGGTATACAATATGAAGAGCCAATAAAGACATCATGGCGTCCACCACGTTGCATCCTCGAGCTGCCGGAGACGAGACACGAAATGATCAGGAATAAATTGAGGATATTAGTCGAGGGTGACGATCCCCCTCCGCCAATAAGGACCTTTCAGCACATGAAGTTTCCTAAAGGCATTCTCCGCGGTTTAGAAGCCAAGGGCATCCAGAAACCGACCCCGATCCAAGTCCAGGGAATGCCAGCTGTGCTGAACGGCAGGGATATGATCGGAATTGCCTTCACCGGCTCCGGGAAGACATTAGTCTTCACTCTCCCAATAATAATGTTCTGTTTGGAGCAAGAAGTGAAAATGCCATTTATACGGAATGAGGGTCCCTACGGCCTGATCATCTGTCCCTCGCGCGAGCTCGCCAAGCAGACCCACGACATAATACAACACTTCATCAGGCACCTCAAGCTAACCGGGAGCCCCGAGATCCGCAGCTGCCTCGCCATCGGAGGCGTAGCCGTCTCCGAATGCATGGAGGTGGTCCAGAAAGGCGTTCACATCATGGTGGCAACGCCGGGGAGGCTGATGGATATGCTGGATAAAAAGATGGTCCGGCTCAACGTCTGCCGGTACCTCTGCATGGACGAAGCTGATCGCATGATCGACATGGGCTTCGAAGAGGACGTGCGAACGATATTCTCTTACTTCGCCGGGCAGAGACAGACGTTACTGTTCAGCGCCACTATGCCGAAGAAGATCCAGAACTTTGCTCGATCAGCTCTGGTCCGTCCGGTGACGGTAAACGTGGGGCGCGCTGGGGCTGCCTCGCTCACCGTCCGCCAGGACCTGGAGCCGGCGCAGCCCGAGGCCCGCACCGTGCAGCTGCTGCACTGCCTGCAGAAGACTCCGCCCCCGACCCTCATATTCGCCGAGAGGAAACAGGACGTGGATGCCGTGCACGAGTACTTGCTGCTGAAAGGAGTGGAAGCCGTCGCCATTCACGGTGGCAAGGACCAGGAGGAGAGGTCTCGGGCAGTCGAAGCCTTCAGGAGGGGGGAAAAGGACGTGCTGGTGGCCACGGATGTCGCTAGCAAAGGGCTCGACTTCGCGAACATCCAGCACGTCATCAACTACGACATGCCGGAGGACATCGAGAACTACGTGCACCGCATCGGGCGCACGGGGCGCGCGGGGGGCGGGGGCGTGGCCACCACGCTGCTGGGCCGCCCCGACCACAGCGTGCTGCGGGACCTGGTGCATCTGCTGCGGGAAGCCGGACAGCGGGTCCCTGGCTTCCTCCTCGACATGCTGGGTGAGCCCGATGTGCCGGCTCCCGCTGGGCAGGGCTGCTCTTACTGCGCCGGTCTGGGACACAGGATCACAGAATGTCCGAAGTTGGAAGCCGTGCAAAATAAGCAAGCATCGAATATCGGCAGAAGAGACTATTTAGCGAACACCGCAGCCGATTATTAA
Protein
MSDVKDDPAPPPPIKRYRREEKSDSDEDVDNYEPYVPIKERKKQQLLKLGRLGLLTAEAAADTKSSSENDHDDETSQEEWGRRYNVSLLDQHSELKRLAEARALSAAEKQAREEEHILESVAQSKALMGVAELAKGIQYEEPIKTSWRPPRCILELPETRHEMIRNKLRILVEGDDPPPPIRTFQHMKFPKGILRGLEAKGIQKPTPIQVQGMPAVLNGRDMIGIAFTGSGKTLVFTLPIIMFCLEQEVKMPFIRNEGPYGLIICPSRELAKQTHDIIQHFIRHLKLTGSPEIRSCLAIGGVAVSECMEVVQKGVHIMVATPGRLMDMLDKKMVRLNVCRYLCMDEADRMIDMGFEEDVRTIFSYFAGQRQTLLFSATMPKKIQNFARSALVRPVTVNVGRAGAASLTVRQDLEPAQPEARTVQLLHCLQKTPPPTLIFAERKQDVDAVHEYLLLKGVEAVAIHGGKDQEERSRAVEAFRRGEKDVLVATDVASKGLDFANIQHVINYDMPEDIENYVHRIGRTGRAGGGGVATTLLGRPDHSVLRDLVHLLREAGQRVPGFLLDMLGEPDVPAPAGQGCSYCAGLGHRITECPKLEAVQNKQASNIGRRDYLANTAADY
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Similarity
Belongs to the DEAD box helicase family. DDX41 subfamily.
Belongs to the DEAD box helicase family.
Belongs to the DEAD box helicase family.
Keywords
ATP-binding
Complete proteome
Helicase
Hydrolase
Metal-binding
Nucleotide-binding
Nucleus
Phosphoprotein
Reference proteome
RNA-binding
Zinc
Zinc-finger
Feature
chain ATP-dependent RNA helicase abstrakt
Uniprot
H9IX16
A0A2H1V5W2
A0A2A4JY37
A0A194Q562
A0A2W1BKQ2
A0A212FAZ8
+ More
A0A088AF35 W8CDH8 A0A0M8ZXY1 A0A1I8P5Z0 Q16P64 A0A034WDB5 A0A0L0CLV5 A0A0A1WKQ1 A0A182GW89 A0A1A9WIF5 A0A1A9XMW9 A0A0L7QYI4 A0A0K8WJ85 A0A1A9UQ66 A0A1B0A8V5 A0A1Q3F9N1 A0A084W1B2 A0A1B0FHE7 A0A1I8MPP3 A0A0M4EP56 A0A154P3L4 B0X6G6 A0A1Y1M9Z8 A0A182YI74 D6X2V8 A0A067RHY6 A0A2A3EV04 Q7PMT8 A0A232EK57 A0A182R2V2 A0A0T6AYJ4 Q9V3C0 A0A182QU54 E2BNJ7 A0A0Q9X443 A0A182WAV1 A0A195FYG6 A0A182SN58 A0A151WMK9 A0A195BPG9 A0A158NVJ4 A0A2J7R1J9 A0A1W4VY25 A0A0P8XZD5 A0A3L8DKI4 E9J702 A0A1B0CS55 F4WLE2 E2AP94 A0A0J7KZD3 A0A195CRK3 A0A0R1DZS8 A0A0Q9X313 A0A0V0G629 A0A224XGP5 A0A1B6FKG0 A0A1B6DQA1 A0A069DVS9 A0A0Q9VZM8 A0A1Y1M6G4 A0A3B0KWD3 A0A336MN12 A0A2H8TYN9 A0A182NN96 I7LAZ0 A0A2S2R6K4 J9JS78 A0A0P4VST1 A0A2M4CTH6 N6U0W9 U4TY85 T1IAT3 A0A0N1ICT6 A0A182MS14 A0A087TXT5 A0A0A9X672 A0A182FLS1 A0A0K8T9H6 A0A2R5LLQ4 A0A131XEK1 L7MM20 A0A131YTH2 A0A023GNX2 A0A224YNP8 A0A131XUY6 A0A1E1XCK6 V4A307 T1IIY9 A0A1J1HGA2 A0A0L8GXL4 B7P3Q1 A0A0N8DCA1 A0A0P4Z3W2 K1QUK3
A0A088AF35 W8CDH8 A0A0M8ZXY1 A0A1I8P5Z0 Q16P64 A0A034WDB5 A0A0L0CLV5 A0A0A1WKQ1 A0A182GW89 A0A1A9WIF5 A0A1A9XMW9 A0A0L7QYI4 A0A0K8WJ85 A0A1A9UQ66 A0A1B0A8V5 A0A1Q3F9N1 A0A084W1B2 A0A1B0FHE7 A0A1I8MPP3 A0A0M4EP56 A0A154P3L4 B0X6G6 A0A1Y1M9Z8 A0A182YI74 D6X2V8 A0A067RHY6 A0A2A3EV04 Q7PMT8 A0A232EK57 A0A182R2V2 A0A0T6AYJ4 Q9V3C0 A0A182QU54 E2BNJ7 A0A0Q9X443 A0A182WAV1 A0A195FYG6 A0A182SN58 A0A151WMK9 A0A195BPG9 A0A158NVJ4 A0A2J7R1J9 A0A1W4VY25 A0A0P8XZD5 A0A3L8DKI4 E9J702 A0A1B0CS55 F4WLE2 E2AP94 A0A0J7KZD3 A0A195CRK3 A0A0R1DZS8 A0A0Q9X313 A0A0V0G629 A0A224XGP5 A0A1B6FKG0 A0A1B6DQA1 A0A069DVS9 A0A0Q9VZM8 A0A1Y1M6G4 A0A3B0KWD3 A0A336MN12 A0A2H8TYN9 A0A182NN96 I7LAZ0 A0A2S2R6K4 J9JS78 A0A0P4VST1 A0A2M4CTH6 N6U0W9 U4TY85 T1IAT3 A0A0N1ICT6 A0A182MS14 A0A087TXT5 A0A0A9X672 A0A182FLS1 A0A0K8T9H6 A0A2R5LLQ4 A0A131XEK1 L7MM20 A0A131YTH2 A0A023GNX2 A0A224YNP8 A0A131XUY6 A0A1E1XCK6 V4A307 T1IIY9 A0A1J1HGA2 A0A0L8GXL4 B7P3Q1 A0A0N8DCA1 A0A0P4Z3W2 K1QUK3
EC Number
3.6.4.13
Pubmed
19121390
26354079
28756777
22118469
24495485
17510324
+ More
25348373 26108605 25830018 26483478 24438588 25315136 28004739 25244985 18362917 19820115 24845553 12364791 14747013 17210077 28648823 10704843 10607561 10731132 12537572 12537569 18327897 20798317 17994087 21347285 30249741 21282665 21719571 17550304 26334808 15632085 27129103 23537049 25401762 28049606 25576852 26830274 28797301 28503490 23254933 22992520
25348373 26108605 25830018 26483478 24438588 25315136 28004739 25244985 18362917 19820115 24845553 12364791 14747013 17210077 28648823 10704843 10607561 10731132 12537572 12537569 18327897 20798317 17994087 21347285 30249741 21282665 21719571 17550304 26334808 15632085 27129103 23537049 25401762 28049606 25576852 26830274 28797301 28503490 23254933 22992520
EMBL
BABH01021230
BABH01021231
ODYU01000851
SOQ36240.1
NWSH01000399
PCG76686.1
+ More
KQ459463 KPJ00499.1 KZ149995 PZC75468.1 AGBW02009404 OWR50899.1 GAMC01001921 JAC04635.1 KQ435821 KOX72336.1 CH477792 EAT36170.1 GAKP01007199 JAC51753.1 JRES01000310 KNC32409.1 GBXI01015324 JAC98967.1 JXUM01092880 KQ564017 KXJ72932.1 KQ414688 KOC63642.1 GDHF01001103 JAI51211.1 GFDL01010803 JAV24242.1 ATLV01019254 KE525266 KFB44006.1 CCAG010022398 CP012526 ALC45822.1 KQ434803 KZC05934.1 DS232412 EDS41449.1 GEZM01041772 GEZM01041767 GEZM01041764 GEZM01041762 JAV80167.1 KQ971372 EFA10652.1 KK852669 KDR18857.1 KZ288185 PBC34901.1 AAAB01008968 EAA13218.5 NNAY01003892 OXU18708.1 LJIG01022542 KRT80027.1 AF212866 AF187729 AE014297 AY051752 AXCN02001903 GL449414 EFN82776.1 CH964251 KRF99725.1 KQ981169 KYN45347.1 KQ982944 KYQ49041.1 KQ976433 KYM87349.1 ADTU01027228 NEVH01008206 PNF34710.1 CH902617 KPU79939.1 QOIP01000007 RLU20954.1 GL768421 EFZ11376.1 AJWK01025650 GL888207 EGI65105.1 GL441481 EFN64767.1 LBMM01001801 KMQ95726.1 KQ977349 KYN03373.1 CM000160 KRK02571.1 CH933806 KRG02432.1 GECL01002944 JAP03180.1 GFTR01007438 JAW08988.1 GECZ01019094 JAS50675.1 GEDC01009446 JAS27852.1 GBGD01000874 JAC88015.1 CH940656 KRF78298.1 GEZM01041770 GEZM01041766 JAV80180.1 OUUW01000014 SPP88318.1 UFQS01001426 UFQT01001426 SSX10839.1 SSX30519.1 GFXV01007562 MBW19367.1 CM000070 EAL28779.1 GGMS01016484 MBY85687.1 ABLF02021400 GDKW01001561 JAI55034.1 GGFL01004459 MBW68637.1 APGK01043999 KB741019 ENN75170.1 KB631645 ERL84963.1 ACPB03005775 KQ459757 KPJ20150.1 AXCM01012325 KK117242 KFM69924.1 GBHO01029284 GBHO01029283 GBHO01029282 GBHO01029281 GBHO01029280 GBHO01029279 JAG14320.1 JAG14321.1 JAG14322.1 JAG14323.1 JAG14324.1 JAG14325.1 GBRD01003632 JAG62189.1 GGLE01006293 MBY10419.1 GEFH01003544 JAP65037.1 GACK01000770 JAA64264.1 GEDV01007121 JAP81436.1 GBBM01000648 JAC34770.1 GFPF01006223 MAA17369.1 GEFM01006415 JAP69381.1 GFAC01002201 JAT96987.1 KB202284 ESO91087.1 JH430212 CVRI01000003 CRK87032.1 KQ420026 KOF81594.1 ABJB010176029 ABJB010372964 ABJB010539317 ABJB010615512 ABJB010696161 ABJB010754891 ABJB010770624 ABJB010999383 ABJB011073380 DS630327 EEC01223.1 GDIP01047839 JAM55876.1 GDIP01219824 JAJ03578.1 JH819154 EKC40522.1
KQ459463 KPJ00499.1 KZ149995 PZC75468.1 AGBW02009404 OWR50899.1 GAMC01001921 JAC04635.1 KQ435821 KOX72336.1 CH477792 EAT36170.1 GAKP01007199 JAC51753.1 JRES01000310 KNC32409.1 GBXI01015324 JAC98967.1 JXUM01092880 KQ564017 KXJ72932.1 KQ414688 KOC63642.1 GDHF01001103 JAI51211.1 GFDL01010803 JAV24242.1 ATLV01019254 KE525266 KFB44006.1 CCAG010022398 CP012526 ALC45822.1 KQ434803 KZC05934.1 DS232412 EDS41449.1 GEZM01041772 GEZM01041767 GEZM01041764 GEZM01041762 JAV80167.1 KQ971372 EFA10652.1 KK852669 KDR18857.1 KZ288185 PBC34901.1 AAAB01008968 EAA13218.5 NNAY01003892 OXU18708.1 LJIG01022542 KRT80027.1 AF212866 AF187729 AE014297 AY051752 AXCN02001903 GL449414 EFN82776.1 CH964251 KRF99725.1 KQ981169 KYN45347.1 KQ982944 KYQ49041.1 KQ976433 KYM87349.1 ADTU01027228 NEVH01008206 PNF34710.1 CH902617 KPU79939.1 QOIP01000007 RLU20954.1 GL768421 EFZ11376.1 AJWK01025650 GL888207 EGI65105.1 GL441481 EFN64767.1 LBMM01001801 KMQ95726.1 KQ977349 KYN03373.1 CM000160 KRK02571.1 CH933806 KRG02432.1 GECL01002944 JAP03180.1 GFTR01007438 JAW08988.1 GECZ01019094 JAS50675.1 GEDC01009446 JAS27852.1 GBGD01000874 JAC88015.1 CH940656 KRF78298.1 GEZM01041770 GEZM01041766 JAV80180.1 OUUW01000014 SPP88318.1 UFQS01001426 UFQT01001426 SSX10839.1 SSX30519.1 GFXV01007562 MBW19367.1 CM000070 EAL28779.1 GGMS01016484 MBY85687.1 ABLF02021400 GDKW01001561 JAI55034.1 GGFL01004459 MBW68637.1 APGK01043999 KB741019 ENN75170.1 KB631645 ERL84963.1 ACPB03005775 KQ459757 KPJ20150.1 AXCM01012325 KK117242 KFM69924.1 GBHO01029284 GBHO01029283 GBHO01029282 GBHO01029281 GBHO01029280 GBHO01029279 JAG14320.1 JAG14321.1 JAG14322.1 JAG14323.1 JAG14324.1 JAG14325.1 GBRD01003632 JAG62189.1 GGLE01006293 MBY10419.1 GEFH01003544 JAP65037.1 GACK01000770 JAA64264.1 GEDV01007121 JAP81436.1 GBBM01000648 JAC34770.1 GFPF01006223 MAA17369.1 GEFM01006415 JAP69381.1 GFAC01002201 JAT96987.1 KB202284 ESO91087.1 JH430212 CVRI01000003 CRK87032.1 KQ420026 KOF81594.1 ABJB010176029 ABJB010372964 ABJB010539317 ABJB010615512 ABJB010696161 ABJB010754891 ABJB010770624 ABJB010999383 ABJB011073380 DS630327 EEC01223.1 GDIP01047839 JAM55876.1 GDIP01219824 JAJ03578.1 JH819154 EKC40522.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000005203
UP000053105
+ More
UP000095300 UP000008820 UP000037069 UP000069940 UP000249989 UP000091820 UP000092443 UP000053825 UP000078200 UP000092445 UP000030765 UP000092444 UP000095301 UP000092553 UP000076502 UP000002320 UP000076408 UP000007266 UP000027135 UP000242457 UP000007062 UP000215335 UP000075900 UP000000803 UP000075886 UP000008237 UP000007798 UP000075920 UP000078541 UP000075901 UP000075809 UP000078540 UP000005205 UP000235965 UP000192221 UP000007801 UP000279307 UP000092461 UP000007755 UP000000311 UP000036403 UP000078542 UP000002282 UP000009192 UP000008792 UP000268350 UP000075884 UP000001819 UP000007819 UP000019118 UP000030742 UP000015103 UP000053240 UP000075883 UP000054359 UP000069272 UP000030746 UP000183832 UP000053454 UP000001555 UP000005408
UP000095300 UP000008820 UP000037069 UP000069940 UP000249989 UP000091820 UP000092443 UP000053825 UP000078200 UP000092445 UP000030765 UP000092444 UP000095301 UP000092553 UP000076502 UP000002320 UP000076408 UP000007266 UP000027135 UP000242457 UP000007062 UP000215335 UP000075900 UP000000803 UP000075886 UP000008237 UP000007798 UP000075920 UP000078541 UP000075901 UP000075809 UP000078540 UP000005205 UP000235965 UP000192221 UP000007801 UP000279307 UP000092461 UP000007755 UP000000311 UP000036403 UP000078542 UP000002282 UP000009192 UP000008792 UP000268350 UP000075884 UP000001819 UP000007819 UP000019118 UP000030742 UP000015103 UP000053240 UP000075883 UP000054359 UP000069272 UP000030746 UP000183832 UP000053454 UP000001555 UP000005408
Interpro
CDD
ProteinModelPortal
H9IX16
A0A2H1V5W2
A0A2A4JY37
A0A194Q562
A0A2W1BKQ2
A0A212FAZ8
+ More
A0A088AF35 W8CDH8 A0A0M8ZXY1 A0A1I8P5Z0 Q16P64 A0A034WDB5 A0A0L0CLV5 A0A0A1WKQ1 A0A182GW89 A0A1A9WIF5 A0A1A9XMW9 A0A0L7QYI4 A0A0K8WJ85 A0A1A9UQ66 A0A1B0A8V5 A0A1Q3F9N1 A0A084W1B2 A0A1B0FHE7 A0A1I8MPP3 A0A0M4EP56 A0A154P3L4 B0X6G6 A0A1Y1M9Z8 A0A182YI74 D6X2V8 A0A067RHY6 A0A2A3EV04 Q7PMT8 A0A232EK57 A0A182R2V2 A0A0T6AYJ4 Q9V3C0 A0A182QU54 E2BNJ7 A0A0Q9X443 A0A182WAV1 A0A195FYG6 A0A182SN58 A0A151WMK9 A0A195BPG9 A0A158NVJ4 A0A2J7R1J9 A0A1W4VY25 A0A0P8XZD5 A0A3L8DKI4 E9J702 A0A1B0CS55 F4WLE2 E2AP94 A0A0J7KZD3 A0A195CRK3 A0A0R1DZS8 A0A0Q9X313 A0A0V0G629 A0A224XGP5 A0A1B6FKG0 A0A1B6DQA1 A0A069DVS9 A0A0Q9VZM8 A0A1Y1M6G4 A0A3B0KWD3 A0A336MN12 A0A2H8TYN9 A0A182NN96 I7LAZ0 A0A2S2R6K4 J9JS78 A0A0P4VST1 A0A2M4CTH6 N6U0W9 U4TY85 T1IAT3 A0A0N1ICT6 A0A182MS14 A0A087TXT5 A0A0A9X672 A0A182FLS1 A0A0K8T9H6 A0A2R5LLQ4 A0A131XEK1 L7MM20 A0A131YTH2 A0A023GNX2 A0A224YNP8 A0A131XUY6 A0A1E1XCK6 V4A307 T1IIY9 A0A1J1HGA2 A0A0L8GXL4 B7P3Q1 A0A0N8DCA1 A0A0P4Z3W2 K1QUK3
A0A088AF35 W8CDH8 A0A0M8ZXY1 A0A1I8P5Z0 Q16P64 A0A034WDB5 A0A0L0CLV5 A0A0A1WKQ1 A0A182GW89 A0A1A9WIF5 A0A1A9XMW9 A0A0L7QYI4 A0A0K8WJ85 A0A1A9UQ66 A0A1B0A8V5 A0A1Q3F9N1 A0A084W1B2 A0A1B0FHE7 A0A1I8MPP3 A0A0M4EP56 A0A154P3L4 B0X6G6 A0A1Y1M9Z8 A0A182YI74 D6X2V8 A0A067RHY6 A0A2A3EV04 Q7PMT8 A0A232EK57 A0A182R2V2 A0A0T6AYJ4 Q9V3C0 A0A182QU54 E2BNJ7 A0A0Q9X443 A0A182WAV1 A0A195FYG6 A0A182SN58 A0A151WMK9 A0A195BPG9 A0A158NVJ4 A0A2J7R1J9 A0A1W4VY25 A0A0P8XZD5 A0A3L8DKI4 E9J702 A0A1B0CS55 F4WLE2 E2AP94 A0A0J7KZD3 A0A195CRK3 A0A0R1DZS8 A0A0Q9X313 A0A0V0G629 A0A224XGP5 A0A1B6FKG0 A0A1B6DQA1 A0A069DVS9 A0A0Q9VZM8 A0A1Y1M6G4 A0A3B0KWD3 A0A336MN12 A0A2H8TYN9 A0A182NN96 I7LAZ0 A0A2S2R6K4 J9JS78 A0A0P4VST1 A0A2M4CTH6 N6U0W9 U4TY85 T1IAT3 A0A0N1ICT6 A0A182MS14 A0A087TXT5 A0A0A9X672 A0A182FLS1 A0A0K8T9H6 A0A2R5LLQ4 A0A131XEK1 L7MM20 A0A131YTH2 A0A023GNX2 A0A224YNP8 A0A131XUY6 A0A1E1XCK6 V4A307 T1IIY9 A0A1J1HGA2 A0A0L8GXL4 B7P3Q1 A0A0N8DCA1 A0A0P4Z3W2 K1QUK3
PDB
5H1Y
E-value=3.21011e-109,
Score=1012
Ontologies
GO
Topology
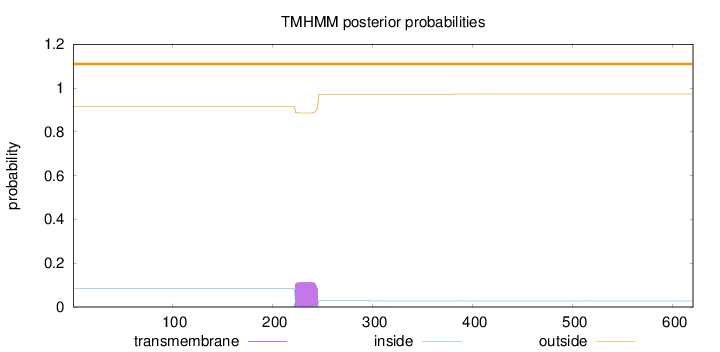
Subcellular location
Nucleus
Length:
620
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.57233000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08555
outside
1 - 620
Population Genetic Test Statistics
Pi
268.261029
Theta
182.184673
Tajima's D
0.94829
CLR
0.642291
CSRT
0.64136793160342
Interpretation
Uncertain
