Gene
KWMTBOMO06221 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001648
Annotation
PREDICTED:_ferrochelatase?_mitochondrial-like_[Bombyx_mori]
Full name
Ferrochelatase
+ More
Ferrochelatase, mitochondrial
Ferrochelatase, mitochondrial
Alternative Name
Heme synthase
Protoheme ferro-lyase
Protoheme ferro-lyase
Location in the cell
Mitochondrial Reliability : 2.264
Sequence
CDS
ATGATGTCTAAAATTCATCGCTTCAATTCCATGATACTACCTCGTTTACCTCCTGCATTCAGCACGGCTTCAGGAACCGCTGTCAAACCTAAGACTGCTATAGTCATGCTGAATATGGGTGGGCCGGAGACCACGGACCAGGTTGGTGACTATCTTCACAGGATAATGACAGACAGAGACATGATCCAGCTTCCGGTGCAGAGTAAACTGGGACCGTGGATAGCCAGTAGAAGGACCGAGGAAGTCAAGAAGAAGTATTCGGAGATTGGTGGAGGATCACCTATATTGAAATGGACAAATAGTCAAGGTAGCTTATTAACGCAGGCCTTAGATCAGATGCTCCCGGAGTCAGCCCCTCATAAGCACTACGTGGCGTTCAGATACGTTCCTCCGTTCACGGAAGACGCCTTCAGGGAGGTGGAAAGCGACGGAGCGGACAGAGTTATCATATTCTCGCAGTATCCTCAGTACAGTTGTGCGACGACTGGTTCTAGTCTGAACGCGATCGCGGATTTCTATAGGAACAGGACGACACCGGCAAACGTAAAGTTTAGTTTGATAGAACGCTGGGCGTCCCACAAGCTGTTGGCGAAGGTCTTCTCCGAGAGGATCAAGGAGAAACTGGACAAATTCGATGAGAAGATCAGGGATGACGTATTGATACTGTTCACGGCGCATAGTCTGCCTTTGAAGGCCGTATCGAGAGGCGACACGTATCCTCACGAGGTGGCCTCCACCGTATCGAGGACCATGGAGGAGCTCGGCGGCCGGAATCCGCACCGGCTGGTCTGGCAGTCGAAGGTGGGCCCCCTGCCCTGGCTCCAGCCCTATACCGACGACGCGATCAAGTTGTACGCGAAGCAAGGAGTCAAGCACCTGATCCTGGTGCCAGTGGCCTTCGTCAACGAACACATCGAGACCCTGCACGAGCTGGACATCGAGTACTGCGACGAGGTAGCCAAGGAGGCCGGGATCCTCCAAATCGAGAGGGCGGAGGCGCCGAACGTGCACCCCACGTTCATCGCGGCGCTGGCGGACGTCGTCGCGGGCCACATCAGCAACGGACCGAGGATCTCGAAGCAGTTCCTGACCAGATGTCCACATTGCGTGAGTCAGAGATGTCTCGCGTCGAAAGAGTTCTTCAAGAGATTGTGTGGCGCCGATCAGAACGTGGATAAAATAGGATCTTAA
Protein
MMSKIHRFNSMILPRLPPAFSTASGTAVKPKTAIVMLNMGGPETTDQVGDYLHRIMTDRDMIQLPVQSKLGPWIASRRTEEVKKKYSEIGGGSPILKWTNSQGSLLTQALDQMLPESAPHKHYVAFRYVPPFTEDAFREVESDGADRVIIFSQYPQYSCATTGSSLNAIADFYRNRTTPANVKFSLIERWASHKLLAKVFSERIKEKLDKFDEKIRDDVLILFTAHSLPLKAVSRGDTYPHEVASTVSRTMEELGGRNPHRLVWQSKVGPLPWLQPYTDDAIKLYAKQGVKHLILVPVAFVNEHIETLHELDIEYCDEVAKEAGILQIERAEAPNVHPTFIAALADVVAGHISNGPRISKQFLTRCPHCVSQRCLASKEFFKRLCGADQNVDKIGS
Summary
Description
Catalyzes the ferrous insertion into protoporphyrin IX.
Catalytic Activity
2 H(+) + heme b = Fe(2+) + protoporphyrin IX
Cofactor
[2Fe-2S] cluster
Similarity
Belongs to the ferrochelatase family.
Keywords
2Fe-2S
Alternative splicing
Complete proteome
Heme biosynthesis
Iron
Iron-sulfur
Lyase
Membrane
Metal-binding
Mitochondrion
Mitochondrion inner membrane
Porphyrin biosynthesis
Reference proteome
Transit peptide
Feature
chain Ferrochelatase
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9IWL6
A0A194Q673
A0A2A4K1G5
A0A0N0PDD9
A0A1I8NIE5
A0A0A1WJI0
+ More
A0A0L0CEU2 A0A1I8Q922 A0A0K8VXJ2 A0A0K8UYX0 W8C3L1 A0A034WXW5 A0A1L8DZM4 A0A1I8JVS6 A0A1L8DZL5 A0A182VEE7 A0A182I916 A0A232EQE6 A0A182UDS9 A0A0M4F540 A0A182LAZ9 Q7QG25 A0A0C9R3D5 K7J6F4 A0A3F2YU13 A0A1B6JBC8 B3LVE6 A0A0K8TL18 A0A182PQQ9 B4K856 A0A182QRZ2 A0A2M3ZGE4 A0A1Q3FF29 B4LY79 B4N956 A0A2M3Z273 A0A182JCM4 A0A340TBF9 T1PJ01 B0WMV1 A0A2Y9D1F0 A0A182HDH9 A0A154P178 A0A2J7RC77 A0A084WR17 Q17A56 A0A182GMR7 A0A2M4ACE0 A0A2J7RC80 B4JTR5 A0A182M850 A0A182S4U9 A0A2M4ACD1 Q967G9 B4PP36 B3P4Z2 A0A2Y9D0I2 A0A3B0K9N3 A0A0R1EBB5 A0A1B0DQV8 A0A1W4VYK9 A0A1W4VZG1 W5JKR8 A0A1B6KI31 B4QTA8 E2BRA4 B4IJ43 E9IFZ2 B4G340 Q297V9 A0A0P4VTD1 A0A336N1E7 Q9V9S8 A0A2M4BR17 E2AB59 C4WVX3 A0A2W1B7B1 A0A1J1J4S4 A0A336MYP4 A0A0A9WFE4 T1DUG1 A0A2S2NQ52 A0A151ICQ7 T1HSU2 A0A151WKA4 A0A195BS91 A0A158NY65 A0A1A9Y931 A0A087ZU01 A0A1B0B6R7 A0A2A3EKZ1 A0A1B0FL77 A0A1B6C2V0 A0A1B6DFR4 A0A182YJM0 A0A310SRK1 A0A067RJX2 A0A1B0AGK9 A0A151ISJ6 F4W9L3
A0A0L0CEU2 A0A1I8Q922 A0A0K8VXJ2 A0A0K8UYX0 W8C3L1 A0A034WXW5 A0A1L8DZM4 A0A1I8JVS6 A0A1L8DZL5 A0A182VEE7 A0A182I916 A0A232EQE6 A0A182UDS9 A0A0M4F540 A0A182LAZ9 Q7QG25 A0A0C9R3D5 K7J6F4 A0A3F2YU13 A0A1B6JBC8 B3LVE6 A0A0K8TL18 A0A182PQQ9 B4K856 A0A182QRZ2 A0A2M3ZGE4 A0A1Q3FF29 B4LY79 B4N956 A0A2M3Z273 A0A182JCM4 A0A340TBF9 T1PJ01 B0WMV1 A0A2Y9D1F0 A0A182HDH9 A0A154P178 A0A2J7RC77 A0A084WR17 Q17A56 A0A182GMR7 A0A2M4ACE0 A0A2J7RC80 B4JTR5 A0A182M850 A0A182S4U9 A0A2M4ACD1 Q967G9 B4PP36 B3P4Z2 A0A2Y9D0I2 A0A3B0K9N3 A0A0R1EBB5 A0A1B0DQV8 A0A1W4VYK9 A0A1W4VZG1 W5JKR8 A0A1B6KI31 B4QTA8 E2BRA4 B4IJ43 E9IFZ2 B4G340 Q297V9 A0A0P4VTD1 A0A336N1E7 Q9V9S8 A0A2M4BR17 E2AB59 C4WVX3 A0A2W1B7B1 A0A1J1J4S4 A0A336MYP4 A0A0A9WFE4 T1DUG1 A0A2S2NQ52 A0A151ICQ7 T1HSU2 A0A151WKA4 A0A195BS91 A0A158NY65 A0A1A9Y931 A0A087ZU01 A0A1B0B6R7 A0A2A3EKZ1 A0A1B0FL77 A0A1B6C2V0 A0A1B6DFR4 A0A182YJM0 A0A310SRK1 A0A067RJX2 A0A1B0AGK9 A0A151ISJ6 F4W9L3
EC Number
4.99.1.1
Pubmed
19121390
26354079
25315136
25830018
26108605
24495485
+ More
25348373 28648823 20966253 12364791 14747013 17210077 20075255 17994087 26369729 26483478 24438588 17510324 15532727 17550304 20920257 23761445 20798317 21282665 15632085 27129103 9712849 10731132 12537572 12537569 28756777 25401762 26823975 21347285 25244985 24845553 21719571
25348373 28648823 20966253 12364791 14747013 17210077 20075255 17994087 26369729 26483478 24438588 17510324 15532727 17550304 20920257 23761445 20798317 21282665 15632085 27129103 9712849 10731132 12537572 12537569 28756777 25401762 26823975 21347285 25244985 24845553 21719571
EMBL
BABH01021223
BABH01021224
KQ459463
KPJ00505.1
NWSH01000270
PCG77866.1
+ More
KQ460393 KPJ15385.1 GBXI01015083 JAC99208.1 JRES01000501 KNC30737.1 GDHF01009074 JAI43240.1 GDHF01020586 JAI31728.1 GAMC01006017 GAMC01006014 JAC00542.1 GAKP01000324 JAC58628.1 GFDF01002207 JAV11877.1 GFDF01002208 JAV11876.1 APCN01003158 NNAY01002784 OXU20568.1 CP012526 ALC46582.1 AAAB01008844 EAA05967.3 GBYB01010777 GBYB01010779 JAG80544.1 JAG80546.1 GECU01011373 JAS96333.1 CH902617 EDV41474.1 GDAI01002531 JAI15072.1 CH933806 EDW15410.2 AXCN02001183 GGFM01006838 MBW27589.1 GFDL01008892 JAV26153.1 CH940650 EDW67967.1 CH964232 EDW81603.1 GGFM01001886 MBW22637.1 KA647893 AFP62522.1 DS232003 EDS31343.1 JXUM01035014 KQ561044 KXJ79910.1 KQ434785 KZC04998.1 NEVH01005885 PNF38444.1 ATLV01025887 KE525402 KFB52661.1 CH477340 EAT43108.1 JXUM01075026 KQ562865 KXJ74971.1 GGFK01005099 MBW38420.1 PNF38442.1 CH916374 EDV91494.1 AXCM01003114 GGFK01005133 MBW38454.1 AY033074 AAK52337.1 CM000160 EDW99272.1 CH954182 EDV52906.1 OUUW01000005 SPP80258.1 KRK04753.1 AJVK01019411 ADMH02001252 ETN63495.1 GEBQ01028865 JAT11112.1 CM000364 EDX15170.1 GL449942 EFN81767.1 CH480846 EDW50996.1 GL762905 EFZ20501.1 CH479179 EDW24235.1 CM000070 EAL28096.2 GDKW01002391 JAI54204.1 UFQT01001903 SSX32118.1 AF076220 AE014297 AY058251 BT001392 BT001878 GGFJ01006282 MBW55423.1 GL438234 EFN69335.1 ABLF02022108 AK341726 BAH72043.1 KZ150251 PZC71819.1 CVRI01000070 CRL07413.1 UFQT01002445 SSX33477.1 GBHO01037508 GBHO01037507 GDHC01010348 GDHC01008234 JAG06096.1 JAG06097.1 JAQ08281.1 JAQ10395.1 GAMD01000064 JAB01527.1 GGMR01006473 MBY19092.1 KQ978004 KYM98194.1 ACPB03016665 KQ983012 KYQ48251.1 KQ976423 KYM88872.1 ADTU01000590 JXJN01009222 KZ288227 PBC31856.1 CCAG010015535 GEDC01029450 JAS07848.1 GEDC01012781 JAS24517.1 KQ761221 OAD57965.1 KK852636 KDR19726.1 KQ981071 KYN09804.1 GL888033 EGI68999.1
KQ460393 KPJ15385.1 GBXI01015083 JAC99208.1 JRES01000501 KNC30737.1 GDHF01009074 JAI43240.1 GDHF01020586 JAI31728.1 GAMC01006017 GAMC01006014 JAC00542.1 GAKP01000324 JAC58628.1 GFDF01002207 JAV11877.1 GFDF01002208 JAV11876.1 APCN01003158 NNAY01002784 OXU20568.1 CP012526 ALC46582.1 AAAB01008844 EAA05967.3 GBYB01010777 GBYB01010779 JAG80544.1 JAG80546.1 GECU01011373 JAS96333.1 CH902617 EDV41474.1 GDAI01002531 JAI15072.1 CH933806 EDW15410.2 AXCN02001183 GGFM01006838 MBW27589.1 GFDL01008892 JAV26153.1 CH940650 EDW67967.1 CH964232 EDW81603.1 GGFM01001886 MBW22637.1 KA647893 AFP62522.1 DS232003 EDS31343.1 JXUM01035014 KQ561044 KXJ79910.1 KQ434785 KZC04998.1 NEVH01005885 PNF38444.1 ATLV01025887 KE525402 KFB52661.1 CH477340 EAT43108.1 JXUM01075026 KQ562865 KXJ74971.1 GGFK01005099 MBW38420.1 PNF38442.1 CH916374 EDV91494.1 AXCM01003114 GGFK01005133 MBW38454.1 AY033074 AAK52337.1 CM000160 EDW99272.1 CH954182 EDV52906.1 OUUW01000005 SPP80258.1 KRK04753.1 AJVK01019411 ADMH02001252 ETN63495.1 GEBQ01028865 JAT11112.1 CM000364 EDX15170.1 GL449942 EFN81767.1 CH480846 EDW50996.1 GL762905 EFZ20501.1 CH479179 EDW24235.1 CM000070 EAL28096.2 GDKW01002391 JAI54204.1 UFQT01001903 SSX32118.1 AF076220 AE014297 AY058251 BT001392 BT001878 GGFJ01006282 MBW55423.1 GL438234 EFN69335.1 ABLF02022108 AK341726 BAH72043.1 KZ150251 PZC71819.1 CVRI01000070 CRL07413.1 UFQT01002445 SSX33477.1 GBHO01037508 GBHO01037507 GDHC01010348 GDHC01008234 JAG06096.1 JAG06097.1 JAQ08281.1 JAQ10395.1 GAMD01000064 JAB01527.1 GGMR01006473 MBY19092.1 KQ978004 KYM98194.1 ACPB03016665 KQ983012 KYQ48251.1 KQ976423 KYM88872.1 ADTU01000590 JXJN01009222 KZ288227 PBC31856.1 CCAG010015535 GEDC01029450 JAS07848.1 GEDC01012781 JAS24517.1 KQ761221 OAD57965.1 KK852636 KDR19726.1 KQ981071 KYN09804.1 GL888033 EGI68999.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000095301
UP000037069
+ More
UP000095300 UP000076407 UP000075903 UP000075840 UP000215335 UP000075902 UP000092553 UP000075882 UP000007062 UP000002358 UP000075881 UP000007801 UP000075885 UP000009192 UP000075886 UP000008792 UP000007798 UP000075880 UP000075920 UP000002320 UP000075884 UP000069940 UP000249989 UP000076502 UP000235965 UP000030765 UP000008820 UP000001070 UP000075883 UP000075900 UP000002282 UP000008711 UP000069272 UP000268350 UP000092462 UP000192221 UP000000673 UP000000304 UP000008237 UP000001292 UP000008744 UP000001819 UP000000803 UP000000311 UP000007819 UP000183832 UP000078542 UP000015103 UP000075809 UP000078540 UP000005205 UP000092443 UP000005203 UP000092460 UP000242457 UP000092444 UP000076408 UP000027135 UP000092445 UP000078492 UP000007755
UP000095300 UP000076407 UP000075903 UP000075840 UP000215335 UP000075902 UP000092553 UP000075882 UP000007062 UP000002358 UP000075881 UP000007801 UP000075885 UP000009192 UP000075886 UP000008792 UP000007798 UP000075880 UP000075920 UP000002320 UP000075884 UP000069940 UP000249989 UP000076502 UP000235965 UP000030765 UP000008820 UP000001070 UP000075883 UP000075900 UP000002282 UP000008711 UP000069272 UP000268350 UP000092462 UP000192221 UP000000673 UP000000304 UP000008237 UP000001292 UP000008744 UP000001819 UP000000803 UP000000311 UP000007819 UP000183832 UP000078542 UP000015103 UP000075809 UP000078540 UP000005205 UP000092443 UP000005203 UP000092460 UP000242457 UP000092444 UP000076408 UP000027135 UP000092445 UP000078492 UP000007755
Interpro
Gene 3D
ProteinModelPortal
H9IWL6
A0A194Q673
A0A2A4K1G5
A0A0N0PDD9
A0A1I8NIE5
A0A0A1WJI0
+ More
A0A0L0CEU2 A0A1I8Q922 A0A0K8VXJ2 A0A0K8UYX0 W8C3L1 A0A034WXW5 A0A1L8DZM4 A0A1I8JVS6 A0A1L8DZL5 A0A182VEE7 A0A182I916 A0A232EQE6 A0A182UDS9 A0A0M4F540 A0A182LAZ9 Q7QG25 A0A0C9R3D5 K7J6F4 A0A3F2YU13 A0A1B6JBC8 B3LVE6 A0A0K8TL18 A0A182PQQ9 B4K856 A0A182QRZ2 A0A2M3ZGE4 A0A1Q3FF29 B4LY79 B4N956 A0A2M3Z273 A0A182JCM4 A0A340TBF9 T1PJ01 B0WMV1 A0A2Y9D1F0 A0A182HDH9 A0A154P178 A0A2J7RC77 A0A084WR17 Q17A56 A0A182GMR7 A0A2M4ACE0 A0A2J7RC80 B4JTR5 A0A182M850 A0A182S4U9 A0A2M4ACD1 Q967G9 B4PP36 B3P4Z2 A0A2Y9D0I2 A0A3B0K9N3 A0A0R1EBB5 A0A1B0DQV8 A0A1W4VYK9 A0A1W4VZG1 W5JKR8 A0A1B6KI31 B4QTA8 E2BRA4 B4IJ43 E9IFZ2 B4G340 Q297V9 A0A0P4VTD1 A0A336N1E7 Q9V9S8 A0A2M4BR17 E2AB59 C4WVX3 A0A2W1B7B1 A0A1J1J4S4 A0A336MYP4 A0A0A9WFE4 T1DUG1 A0A2S2NQ52 A0A151ICQ7 T1HSU2 A0A151WKA4 A0A195BS91 A0A158NY65 A0A1A9Y931 A0A087ZU01 A0A1B0B6R7 A0A2A3EKZ1 A0A1B0FL77 A0A1B6C2V0 A0A1B6DFR4 A0A182YJM0 A0A310SRK1 A0A067RJX2 A0A1B0AGK9 A0A151ISJ6 F4W9L3
A0A0L0CEU2 A0A1I8Q922 A0A0K8VXJ2 A0A0K8UYX0 W8C3L1 A0A034WXW5 A0A1L8DZM4 A0A1I8JVS6 A0A1L8DZL5 A0A182VEE7 A0A182I916 A0A232EQE6 A0A182UDS9 A0A0M4F540 A0A182LAZ9 Q7QG25 A0A0C9R3D5 K7J6F4 A0A3F2YU13 A0A1B6JBC8 B3LVE6 A0A0K8TL18 A0A182PQQ9 B4K856 A0A182QRZ2 A0A2M3ZGE4 A0A1Q3FF29 B4LY79 B4N956 A0A2M3Z273 A0A182JCM4 A0A340TBF9 T1PJ01 B0WMV1 A0A2Y9D1F0 A0A182HDH9 A0A154P178 A0A2J7RC77 A0A084WR17 Q17A56 A0A182GMR7 A0A2M4ACE0 A0A2J7RC80 B4JTR5 A0A182M850 A0A182S4U9 A0A2M4ACD1 Q967G9 B4PP36 B3P4Z2 A0A2Y9D0I2 A0A3B0K9N3 A0A0R1EBB5 A0A1B0DQV8 A0A1W4VYK9 A0A1W4VZG1 W5JKR8 A0A1B6KI31 B4QTA8 E2BRA4 B4IJ43 E9IFZ2 B4G340 Q297V9 A0A0P4VTD1 A0A336N1E7 Q9V9S8 A0A2M4BR17 E2AB59 C4WVX3 A0A2W1B7B1 A0A1J1J4S4 A0A336MYP4 A0A0A9WFE4 T1DUG1 A0A2S2NQ52 A0A151ICQ7 T1HSU2 A0A151WKA4 A0A195BS91 A0A158NY65 A0A1A9Y931 A0A087ZU01 A0A1B0B6R7 A0A2A3EKZ1 A0A1B0FL77 A0A1B6C2V0 A0A1B6DFR4 A0A182YJM0 A0A310SRK1 A0A067RJX2 A0A1B0AGK9 A0A151ISJ6 F4W9L3
PDB
2PO7
E-value=1.25143e-108,
Score=1005
Ontologies
PATHWAY
GO
PANTHER
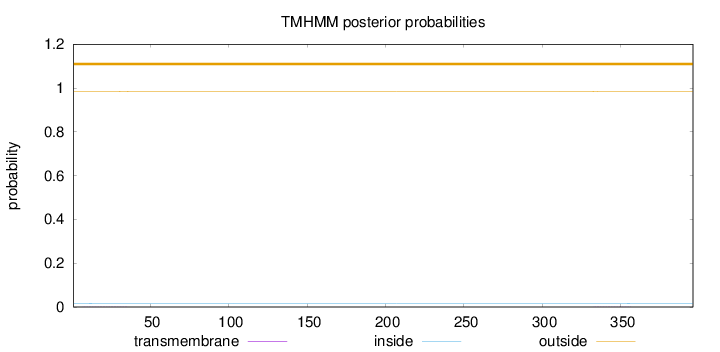
Topology
Subcellular location
Mitochondrion inner membrane
Length:
396
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03744
Exp number, first 60 AAs:
0.01922
Total prob of N-in:
0.01601
outside
1 - 396
Population Genetic Test Statistics
Pi
244.497541
Theta
169.302214
Tajima's D
1.562248
CLR
0.272688
CSRT
0.798410079496025
Interpretation
Uncertain
