Gene
KWMTBOMO06218
Pre Gene Modal
BGIBMGA001650
Annotation
PREDICTED:_leishmanolysin-like_peptidase_isoform_X2_[Bombyx_mori]
Full name
Leishmanolysin-like peptidase
+ More
Phosphoglycerate mutase
Phosphoglycerate mutase
Alternative Name
Invadolysin
Location in the cell
Extracellular Reliability : 2.63
Sequence
CDS
ATGTGTGGAGAGGTTCGTGTACCAGAGGAGCATTTGAATCCTTGCTATGTCTGTAATAGCACCGGCCACGACTGCCGGGCGCTACCTCGCTACCAGCCCCCTCCGGATGATGAACCAGCTACAGAAGTCAGCGAGAACAATCTCGACGAACGAAGCGATGTTATCGATGAGGATTCGGAGCCGGTCGGCGTGAAGGATACTGACTTCATGCTCTACGTCAGCGCCGTGGAGACGGAGAGATGCCGGCGGGGTCTCACGGTGGCCTACGCGTCACACTGCCAACAGGAGTCGGCTTTGGACAGGCCGGTGGCTGGTCACGCGAATTTCTGTCCGGCCGAACTGTCGACCAAATACCGCGACTTACCGTCGGTCCTTTCGACCGTCAAACACGAGATGCTGCACGCTCTGGGCTTCAGTGTGTCTCTGTTCGCGTTTTACAGAGACGACAACGGCGAGCCCCTGACCGAAAGAAGACCTGACACCGGGAATCCACCATTGGATGAGGAGCTGCAGATACACAAGTGGTCTGATAGAGTGGTCAGGAACGTGACGCGAAAGAATTGGATGATCAGAGGTGGATATATGGAGAGGACCTTCCACATGATGGTCACTCCGAGGGTTGTTAAAGAGGTACGCGAACACTTCAACTGCTCAGAGCTGGAGGGGGCAGAGTTGGAGGATCAGGGTGGCGACGGCACCGCCATGACCCACTGGGAGAAGAGAGTCTTTGAGAACGAGGCCATGACCGGCACCCACACCCAGAACTCGGTCTTCAGCAGGATAACCCTCGCGATGATGGAGGACACCGGCTGGTACAGGGCGGACTACTCCCACGCGACACCCCTGGACTGGGGGAAGGGACTGGGCTGTAAATTCGCCATGTCCTCGTGCAAGCAGTGGATGAACCTGCAAAGACGCAGAAATCCGGCGCCGTTTTGCGAGAGGATAAAGGGGAATCCCTTGAGGACAGAGTGCTCCCCGAGGAGGTCGGCAGTCGTCCTCTGCAACCTGGTCCGCCACGACAACCTACTGCCGAGAGCCTACCAGCATTTCGACATACTGCCCAACGTTCCACCAGGGCAGGAAGCGTACTATGGCGGCTCCGTGTCCCTCGCGGACTACTGCCCCTACCTTCAGGAGTTCACTTGGAGACACAAGAGCGTACTCATCAGGGGAAGTCGATGCTCCTACGAGGAGAATACTCCGAAGATCGATCTGAACTTCGCCCTTGAGAACTACGGGCAGCACTCGAAGTGTTTCGAGCACAGCGACAAGGTCTGGGAGCAGAAGTCATGCAGACAGATCCGCGAATGGCAACACTGGGGCTCCGGTTGCTACAAATACAAATGCGACAGCGGTCGTCTGCATATCGTGGTGGGTAATTACACGTACACGTGCTTTCACGCCGGTCAACTGCTGCACATAAGGATCATAAAGAACGGCTGGCTTCACAGGGGGGGCGTGGTCTGTCCCCCCTGCAGGCAAGTGTGCGGCGCTGAGTTCGCTGCGAGAAGCGAATACTGTAAGCCGGGTGAGGAGCCTCTACCCCCGAATTTGTATCCCAACGATTTTCTAGCTTGTAGAGCGTCGGTTATCCGACCCGCCATCTTGTGGACGGCGGCCATCTTGTATTCGCTGTCATAA
Protein
MCGEVRVPEEHLNPCYVCNSTGHDCRALPRYQPPPDDEPATEVSENNLDERSDVIDEDSEPVGVKDTDFMLYVSAVETERCRRGLTVAYASHCQQESALDRPVAGHANFCPAELSTKYRDLPSVLSTVKHEMLHALGFSVSLFAFYRDDNGEPLTERRPDTGNPPLDEELQIHKWSDRVVRNVTRKNWMIRGGYMERTFHMMVTPRVVKEVREHFNCSELEGAELEDQGGDGTAMTHWEKRVFENEAMTGTHTQNSVFSRITLAMMEDTGWYRADYSHATPLDWGKGLGCKFAMSSCKQWMNLQRRRNPAPFCERIKGNPLRTECSPRRSAVVLCNLVRHDNLLPRAYQHFDILPNVPPGQEAYYGGSVSLADYCPYLQEFTWRHKSVLIRGSRCSYEENTPKIDLNFALENYGQHSKCFEHSDKVWEQKSCRQIREWQHWGSGCYKYKCDSGRLHIVVGNYTYTCFHAGQLLHIRIIKNGWLHRGGVVCPPCRQVCGAEFAARSEYCKPGEEPLPPNLYPNDFLACRASVIRPAILWTAAILYSLS
Summary
Description
Essential for the coordination of mitotic progression, and also plays a role in cell migration.
Catalytic Activity
(2R)-2-phosphoglycerate = 3-phospho-D-glycerate
3-phospho-D-glyceroyl phosphate = (2R)-2,3-bisphosphoglycerate + H(+)
3-phospho-D-glyceroyl phosphate = (2R)-2,3-bisphosphoglycerate + H(+)
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M8 family.
Belongs to the phosphoglycerate mutase family. BPG-dependent PGAM subfamily.
Belongs to the phosphoglycerate mutase family. BPG-dependent PGAM subfamily.
Keywords
Cell cycle
Cell division
Complete proteome
Cytoplasm
Hydrolase
Metal-binding
Metalloprotease
Mitosis
Protease
Reference proteome
Zinc
Feature
chain Leishmanolysin-like peptidase
Uniprot
H9IWL8
A0A2A4JF72
A0A194Q4N7
A0A1S4FEB7
Q175B5
B4K792
+ More
E2AB53 A0A1Q3FUE2 A0A1B0C987 A0A1W4UK25 A0A026VZ23 B4QVA8 A0A3L8DZB7 B3P4K5 B4PKL9 B4M424 Q9VH19 A0A182GDL9 B4NIZ8 E2BRA8 W8ACI8 B4G6G5 A0A0R3NLH2 Q29AK2 A0A1Y1MW84 B3LW16 A0A1W4W9F5 B4JSJ0 A0A1I8N2V8 A0A3B0K087 A0A182FPP9 A0A3B0KPD3 A0A151WK81 A0A195FGG9 A0A0A1WGE6 A0A151ISG8 A0A0M3QY80 T1PIR7 F4W9L1 A0A195BRN4 D6W703 A0A0A1XPI5 A0A2M4CJJ6 W5JHV0 A0A336M4P9 A0A0K8VNN0 A0A084VEG8 A0A0C9QQZ6 A0A0K8ULK9 A0A034VRT6 A0A182JPW5 A0A182HU12 A0A182WRR7 A0A034VV80 A0A0M9ADB2 A0A182UIU6 N6UDW5 A0A151ICR6 A0A182V1P8 A0A182JMK7 Q7QD48 A0A182P6M3 A0A0L0CH49 A0A0L7QNR1 A0A182RNC1 E0VYP7 A0A1B6E3E1 A0A154NZ92 A0A182XWB9 A0A2P8XF71 A0A182VYK7 A0A182MA94 A0A182NDX6 A0A1I8P0D7 A0A1J1HGT9 A0A310SNH9 A0A1B6FAY9 A0A1A9Y4J7 K7J267 A0A1B0AHG5 A0A1B0GDZ8 A0A1A9VM31 A0A232FMG7 A0A2A3EJE5 A0A067QUB0 A0A0T6AVU7 A0A158NCB3 A0A1A9W2N6 A0A2S2Q2P6 A0A0J7KM73 A0A2H8TVJ3 A0A2S2PPE2 A0A182LG15 J9K1Y3 A0A1B6C3F0 B0WA37 A0A3Q0J9U1 A0A1D2NG41 E9H0K4 A0A0A9WRW0
E2AB53 A0A1Q3FUE2 A0A1B0C987 A0A1W4UK25 A0A026VZ23 B4QVA8 A0A3L8DZB7 B3P4K5 B4PKL9 B4M424 Q9VH19 A0A182GDL9 B4NIZ8 E2BRA8 W8ACI8 B4G6G5 A0A0R3NLH2 Q29AK2 A0A1Y1MW84 B3LW16 A0A1W4W9F5 B4JSJ0 A0A1I8N2V8 A0A3B0K087 A0A182FPP9 A0A3B0KPD3 A0A151WK81 A0A195FGG9 A0A0A1WGE6 A0A151ISG8 A0A0M3QY80 T1PIR7 F4W9L1 A0A195BRN4 D6W703 A0A0A1XPI5 A0A2M4CJJ6 W5JHV0 A0A336M4P9 A0A0K8VNN0 A0A084VEG8 A0A0C9QQZ6 A0A0K8ULK9 A0A034VRT6 A0A182JPW5 A0A182HU12 A0A182WRR7 A0A034VV80 A0A0M9ADB2 A0A182UIU6 N6UDW5 A0A151ICR6 A0A182V1P8 A0A182JMK7 Q7QD48 A0A182P6M3 A0A0L0CH49 A0A0L7QNR1 A0A182RNC1 E0VYP7 A0A1B6E3E1 A0A154NZ92 A0A182XWB9 A0A2P8XF71 A0A182VYK7 A0A182MA94 A0A182NDX6 A0A1I8P0D7 A0A1J1HGT9 A0A310SNH9 A0A1B6FAY9 A0A1A9Y4J7 K7J267 A0A1B0AHG5 A0A1B0GDZ8 A0A1A9VM31 A0A232FMG7 A0A2A3EJE5 A0A067QUB0 A0A0T6AVU7 A0A158NCB3 A0A1A9W2N6 A0A2S2Q2P6 A0A0J7KM73 A0A2H8TVJ3 A0A2S2PPE2 A0A182LG15 J9K1Y3 A0A1B6C3F0 B0WA37 A0A3Q0J9U1 A0A1D2NG41 E9H0K4 A0A0A9WRW0
EC Number
3.4.24.-
5.4.2.11
5.4.2.4
5.4.2.11
5.4.2.4
Pubmed
19121390
26354079
17510324
17994087
18057021
20798317
+ More
24508170 30249741 17550304 15557119 10731132 12537572 12537569 26483478 24495485 15632085 28004739 25315136 25830018 21719571 18362917 19820115 20920257 23761445 24438588 25348373 23537049 12364791 14747013 17210077 26108605 20566863 25244985 29403074 20075255 28648823 24845553 21347285 20966253 27289101 21292972 25401762
24508170 30249741 17550304 15557119 10731132 12537572 12537569 26483478 24495485 15632085 28004739 25315136 25830018 21719571 18362917 19820115 20920257 23761445 24438588 25348373 23537049 12364791 14747013 17210077 26108605 20566863 25244985 29403074 20075255 28648823 24845553 21347285 20966253 27289101 21292972 25401762
EMBL
BABH01021211
NWSH01001683
PCG70426.1
KQ459463
KPJ00507.1
CH477403
+ More
EAT41672.1 CH933806 EDW16405.1 KRG02098.1 GL438234 EFN69329.1 GFDL01003804 JAV31241.1 AJWK01002053 AJWK01002054 AJWK01002055 AJWK01002056 AJWK01002057 AJWK01002058 AJWK01002059 KK107555 EZA49022.1 CM000364 EDX13514.1 QOIP01000002 RLU25465.1 CH954181 EDV49658.1 CM000160 EDW96778.1 CH940652 EDW59385.1 AF234630 AE014297 BT001861 JXUM01056215 JXUM01056216 JXUM01056217 KQ561901 KXJ77219.1 CH964272 EDW84900.1 GL449942 EFN81771.1 GAMC01020945 JAB85610.1 CH479179 EDW23884.1 CM000070 KRS99967.1 GEZM01025001 JAV87647.1 CH902617 EDV41549.1 CH916373 EDV94730.1 OUUW01000013 SPP87725.1 SPP87726.1 KQ983012 KYQ48246.1 KQ981606 KYN39476.1 GBXI01016198 JAC98093.1 KQ981071 KYN09812.1 CP012526 ALC47213.1 KA648666 AFP63295.1 GL888033 EGI68997.1 KQ976423 KYM88879.1 KQ971307 EFA11490.2 GBXI01001460 JAD12832.1 GGFL01001237 MBW65415.1 ADMH02001205 ETN63696.1 UFQS01000554 UFQT01000554 SSX04872.1 SSX25235.1 GDHF01020896 GDHF01011827 JAI31418.1 JAI40487.1 ATLV01012277 KE524778 KFB36362.1 GBYB01006149 JAG75916.1 GDHF01024715 JAI27599.1 GAKP01012926 GAKP01012921 JAC46026.1 APCN01000599 GAKP01012925 JAC46027.1 KQ435694 KOX80933.1 APGK01039132 APGK01039133 APGK01039134 KB740967 ENN76852.1 KQ978004 KYM98201.1 AAAB01008859 EAA07618.4 JRES01000409 KNC31527.1 KQ414845 KOC60260.1 DS235845 EEB18503.1 GEDC01004867 JAS32431.1 KQ434785 KZC04996.1 PYGN01002373 PSN30625.1 AXCM01002121 CVRI01000003 CRK87091.1 KQ762805 OAD55473.1 GECZ01022417 JAS47352.1 CCAG010012076 NNAY01000024 OXU31845.1 KZ288227 PBC31857.1 KK853419 KDR07722.1 LJIG01022716 KRT79085.1 ADTU01000559 ADTU01000560 ADTU01000561 ADTU01000562 GGMS01002698 MBY71901.1 LBMM01005602 KMQ91392.1 GFXV01006016 MBW17821.1 GGMR01018722 MBY31341.1 ABLF02039931 ABLF02039936 GEDC01029281 JAS08017.1 DS231868 EDS40770.1 LJIJ01000051 ODN04209.1 GL732581 EFX74736.1 GBHO01032422 JAG11182.1
EAT41672.1 CH933806 EDW16405.1 KRG02098.1 GL438234 EFN69329.1 GFDL01003804 JAV31241.1 AJWK01002053 AJWK01002054 AJWK01002055 AJWK01002056 AJWK01002057 AJWK01002058 AJWK01002059 KK107555 EZA49022.1 CM000364 EDX13514.1 QOIP01000002 RLU25465.1 CH954181 EDV49658.1 CM000160 EDW96778.1 CH940652 EDW59385.1 AF234630 AE014297 BT001861 JXUM01056215 JXUM01056216 JXUM01056217 KQ561901 KXJ77219.1 CH964272 EDW84900.1 GL449942 EFN81771.1 GAMC01020945 JAB85610.1 CH479179 EDW23884.1 CM000070 KRS99967.1 GEZM01025001 JAV87647.1 CH902617 EDV41549.1 CH916373 EDV94730.1 OUUW01000013 SPP87725.1 SPP87726.1 KQ983012 KYQ48246.1 KQ981606 KYN39476.1 GBXI01016198 JAC98093.1 KQ981071 KYN09812.1 CP012526 ALC47213.1 KA648666 AFP63295.1 GL888033 EGI68997.1 KQ976423 KYM88879.1 KQ971307 EFA11490.2 GBXI01001460 JAD12832.1 GGFL01001237 MBW65415.1 ADMH02001205 ETN63696.1 UFQS01000554 UFQT01000554 SSX04872.1 SSX25235.1 GDHF01020896 GDHF01011827 JAI31418.1 JAI40487.1 ATLV01012277 KE524778 KFB36362.1 GBYB01006149 JAG75916.1 GDHF01024715 JAI27599.1 GAKP01012926 GAKP01012921 JAC46026.1 APCN01000599 GAKP01012925 JAC46027.1 KQ435694 KOX80933.1 APGK01039132 APGK01039133 APGK01039134 KB740967 ENN76852.1 KQ978004 KYM98201.1 AAAB01008859 EAA07618.4 JRES01000409 KNC31527.1 KQ414845 KOC60260.1 DS235845 EEB18503.1 GEDC01004867 JAS32431.1 KQ434785 KZC04996.1 PYGN01002373 PSN30625.1 AXCM01002121 CVRI01000003 CRK87091.1 KQ762805 OAD55473.1 GECZ01022417 JAS47352.1 CCAG010012076 NNAY01000024 OXU31845.1 KZ288227 PBC31857.1 KK853419 KDR07722.1 LJIG01022716 KRT79085.1 ADTU01000559 ADTU01000560 ADTU01000561 ADTU01000562 GGMS01002698 MBY71901.1 LBMM01005602 KMQ91392.1 GFXV01006016 MBW17821.1 GGMR01018722 MBY31341.1 ABLF02039931 ABLF02039936 GEDC01029281 JAS08017.1 DS231868 EDS40770.1 LJIJ01000051 ODN04209.1 GL732581 EFX74736.1 GBHO01032422 JAG11182.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000008820
UP000009192
UP000000311
+ More
UP000092461 UP000192221 UP000053097 UP000000304 UP000279307 UP000008711 UP000002282 UP000008792 UP000000803 UP000069940 UP000249989 UP000007798 UP000008237 UP000008744 UP000001819 UP000007801 UP000192223 UP000001070 UP000095301 UP000268350 UP000069272 UP000075809 UP000078541 UP000078492 UP000092553 UP000007755 UP000078540 UP000007266 UP000000673 UP000030765 UP000075881 UP000075840 UP000076407 UP000053105 UP000075902 UP000019118 UP000078542 UP000075903 UP000075880 UP000007062 UP000075885 UP000037069 UP000053825 UP000075900 UP000009046 UP000076502 UP000076408 UP000245037 UP000075920 UP000075883 UP000075884 UP000095300 UP000183832 UP000092443 UP000002358 UP000092445 UP000092444 UP000078200 UP000215335 UP000242457 UP000027135 UP000005205 UP000091820 UP000036403 UP000075882 UP000007819 UP000002320 UP000079169 UP000094527 UP000000305
UP000092461 UP000192221 UP000053097 UP000000304 UP000279307 UP000008711 UP000002282 UP000008792 UP000000803 UP000069940 UP000249989 UP000007798 UP000008237 UP000008744 UP000001819 UP000007801 UP000192223 UP000001070 UP000095301 UP000268350 UP000069272 UP000075809 UP000078541 UP000078492 UP000092553 UP000007755 UP000078540 UP000007266 UP000000673 UP000030765 UP000075881 UP000075840 UP000076407 UP000053105 UP000075902 UP000019118 UP000078542 UP000075903 UP000075880 UP000007062 UP000075885 UP000037069 UP000053825 UP000075900 UP000009046 UP000076502 UP000076408 UP000245037 UP000075920 UP000075883 UP000075884 UP000095300 UP000183832 UP000092443 UP000002358 UP000092445 UP000092444 UP000078200 UP000215335 UP000242457 UP000027135 UP000005205 UP000091820 UP000036403 UP000075882 UP000007819 UP000002320 UP000079169 UP000094527 UP000000305
Interpro
SUPFAM
SSF53254
SSF53254
Gene 3D
ProteinModelPortal
H9IWL8
A0A2A4JF72
A0A194Q4N7
A0A1S4FEB7
Q175B5
B4K792
+ More
E2AB53 A0A1Q3FUE2 A0A1B0C987 A0A1W4UK25 A0A026VZ23 B4QVA8 A0A3L8DZB7 B3P4K5 B4PKL9 B4M424 Q9VH19 A0A182GDL9 B4NIZ8 E2BRA8 W8ACI8 B4G6G5 A0A0R3NLH2 Q29AK2 A0A1Y1MW84 B3LW16 A0A1W4W9F5 B4JSJ0 A0A1I8N2V8 A0A3B0K087 A0A182FPP9 A0A3B0KPD3 A0A151WK81 A0A195FGG9 A0A0A1WGE6 A0A151ISG8 A0A0M3QY80 T1PIR7 F4W9L1 A0A195BRN4 D6W703 A0A0A1XPI5 A0A2M4CJJ6 W5JHV0 A0A336M4P9 A0A0K8VNN0 A0A084VEG8 A0A0C9QQZ6 A0A0K8ULK9 A0A034VRT6 A0A182JPW5 A0A182HU12 A0A182WRR7 A0A034VV80 A0A0M9ADB2 A0A182UIU6 N6UDW5 A0A151ICR6 A0A182V1P8 A0A182JMK7 Q7QD48 A0A182P6M3 A0A0L0CH49 A0A0L7QNR1 A0A182RNC1 E0VYP7 A0A1B6E3E1 A0A154NZ92 A0A182XWB9 A0A2P8XF71 A0A182VYK7 A0A182MA94 A0A182NDX6 A0A1I8P0D7 A0A1J1HGT9 A0A310SNH9 A0A1B6FAY9 A0A1A9Y4J7 K7J267 A0A1B0AHG5 A0A1B0GDZ8 A0A1A9VM31 A0A232FMG7 A0A2A3EJE5 A0A067QUB0 A0A0T6AVU7 A0A158NCB3 A0A1A9W2N6 A0A2S2Q2P6 A0A0J7KM73 A0A2H8TVJ3 A0A2S2PPE2 A0A182LG15 J9K1Y3 A0A1B6C3F0 B0WA37 A0A3Q0J9U1 A0A1D2NG41 E9H0K4 A0A0A9WRW0
E2AB53 A0A1Q3FUE2 A0A1B0C987 A0A1W4UK25 A0A026VZ23 B4QVA8 A0A3L8DZB7 B3P4K5 B4PKL9 B4M424 Q9VH19 A0A182GDL9 B4NIZ8 E2BRA8 W8ACI8 B4G6G5 A0A0R3NLH2 Q29AK2 A0A1Y1MW84 B3LW16 A0A1W4W9F5 B4JSJ0 A0A1I8N2V8 A0A3B0K087 A0A182FPP9 A0A3B0KPD3 A0A151WK81 A0A195FGG9 A0A0A1WGE6 A0A151ISG8 A0A0M3QY80 T1PIR7 F4W9L1 A0A195BRN4 D6W703 A0A0A1XPI5 A0A2M4CJJ6 W5JHV0 A0A336M4P9 A0A0K8VNN0 A0A084VEG8 A0A0C9QQZ6 A0A0K8ULK9 A0A034VRT6 A0A182JPW5 A0A182HU12 A0A182WRR7 A0A034VV80 A0A0M9ADB2 A0A182UIU6 N6UDW5 A0A151ICR6 A0A182V1P8 A0A182JMK7 Q7QD48 A0A182P6M3 A0A0L0CH49 A0A0L7QNR1 A0A182RNC1 E0VYP7 A0A1B6E3E1 A0A154NZ92 A0A182XWB9 A0A2P8XF71 A0A182VYK7 A0A182MA94 A0A182NDX6 A0A1I8P0D7 A0A1J1HGT9 A0A310SNH9 A0A1B6FAY9 A0A1A9Y4J7 K7J267 A0A1B0AHG5 A0A1B0GDZ8 A0A1A9VM31 A0A232FMG7 A0A2A3EJE5 A0A067QUB0 A0A0T6AVU7 A0A158NCB3 A0A1A9W2N6 A0A2S2Q2P6 A0A0J7KM73 A0A2H8TVJ3 A0A2S2PPE2 A0A182LG15 J9K1Y3 A0A1B6C3F0 B0WA37 A0A3Q0J9U1 A0A1D2NG41 E9H0K4 A0A0A9WRW0
PDB
1LML
E-value=5.09322e-27,
Score=302
Ontologies
GO
GO:0016020
GO:0004222
GO:0007155
GO:0016021
GO:0007052
GO:0008406
GO:0007076
GO:0007100
GO:0005737
GO:0008354
GO:0007444
GO:1902769
GO:0007420
GO:0051298
GO:0006338
GO:0019915
GO:0045842
GO:0022900
GO:0031252
GO:0008233
GO:0046872
GO:0051301
GO:0004619
GO:0006096
GO:0004082
GO:0006508
GO:0022857
GO:0004559
GO:0006013
GO:0015923
GO:0005634
GO:0006260
Topology
Subcellular location
Cytoplasm
Found in ring-like structures resembling invadopodia. In migrating cells it relocalizes from internal structures to the leading edge of cells. With evidence from 3 publications.
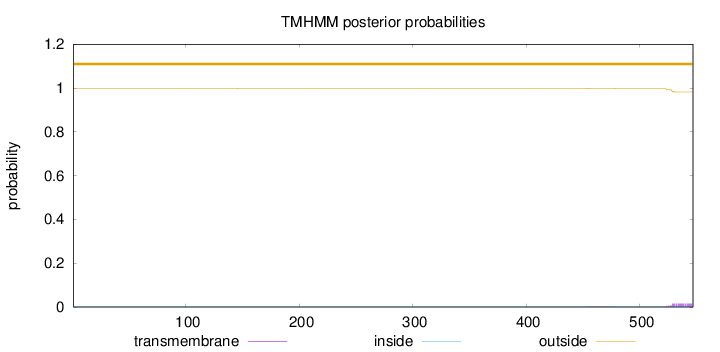
Length:
547
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.35128
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00131
outside
1 - 547
Population Genetic Test Statistics
Pi
209.093649
Theta
152.028559
Tajima's D
1.32481
CLR
10.775044
CSRT
0.744112794360282
Interpretation
Uncertain
