Gene
KWMTBOMO06216 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001793
Annotation
PREDICTED:_sericin_1-like_isoform_X3_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.426
Sequence
CDS
ATGCGTTTCGTTCTGTGCTGCACTTTGATTGCGTTGGCTGCGCTCAGCGTAAAAGCTTTCGGTCACCACCCCGGCAATCGAGATACAGTCGAAGTCAAAAACCGAAAGTACAATGCAGCTAGCAGTGAAAGCTCTTACCTCAACAAAGATAATGATTCGATAAGTGCCGGAGCGCACCGGGCCAAGTCCGTAGAGCAGAGTCAGGATAAAAGCAAATATACATCTGGTCCAGAAGGCGTGTCGTACAGCGGAAGGTCTCAGAACTATAAAGATTCCAAGCAAGCTTATGCCGATTATCACAGCGATCCGAACGGCGGATCTGCTTCTGCCGGACAATCTCGCGACAGCAGCCTGAGAGAGAGAAACGTACATTACGTCTCTGACGGTGAAGCAGTGGCCGCTTCCAGTGACGCTCGCGATGAAAACCGATCCGCCCAACAGAATGCTCAGGCCAATTGGAACGCTGACGGTTCTTACGGAGTTAGCGCTGATCGAAGTGGTTCCGCTAGTTCTAGACGCCGCCAAGCCAATTACTACTCCGATAAAGACATCACTGCTGCTTCTAAAGACGATTCACGTGCAGATTCTTCTAGGAGAAGCAATGCCTATTACAACAGAGATAGTGACGGCTCAGAATCCGCTGGATTAAGTGACCGTAGAGCTTCTTCCTCGAAAAATGATAATGTATTTGTTTACCGCACTAAGGATTCTATTGGAGGACAAGCGAAATCTTCAAGATCATCTCATTCACAAGAGAGCGACGCTTATTATAACTCCAGTCCGGATGGAAGCTACAACGCTGGTACGCGAGATAGTTCAATTTCTAACAAAAAGAAGGCGAGCTCTACCATCTACGCTGATAAAGATCAAATACGCGCCGCGAATGATCGTTATTCTTCGAAACAGTTAAAACAGAGCAGCGCTCAAATCTCCTCCGGGCCAGAGGGCACCTCTGTAAGCAGTAAGGATAGGCAGTACTCGAACGACAAACGCAGCAAATCTGATGCGTACGTCGGACGGGACGGCACCGTTGCTTACTCAAACAAGGACAGCGAAAAGACCTCACGACAAAGTAATACGAACTATGCCGACCAAAACTCCGTTCGCTCTGACTCTGCCGCTTCGGACCAGACCAGCAACAGTTACGACAAGGGCTACAGTGATAAAAATACAGTTGCCCATAGCTCTGGTAGTAGGGGCAGTCAGAATCAGAAATCGTCGAGCTACCGCGCTGACAAGGACGGTTTTTCCTCCAGTACGAATACTGAAAAATCCAGATTTAGTTCTTCGAATAGCGTCGTAGAAACTTCAGATGGAACTTCTGCTAGTCGCGAATCATCAGCGGAGGATACCAAATCATCCAATAGTAACGTTCAGAGCGATGAAACAGGCGAAGAAGAGGAATTGTTCGATGTTGTATCTTACCAGAAAATTGAAGATGGCAAGCCTGTAATCATAATGAAAGTTATACCAGTCGAGAAATCCGCGTCCCAATCAAGTTCTTCGCGGTCATCTCAGGAGTCTGCAAGCTATAGCAGCAGCAGCAGTTCATCGACAGAAGAATCCTCATCCTCGAGCTCTAGGGCTGCTTCATCAACCGACGCTTCTAGCAACACTGATTCAAACTCAAACAGCGCGGGATCCAGTACATCCGGCGGTAGCAGCACTTATGGATACAGTTCCAACAGTCGTGATGGAAGTGTATCGACCACCGGCAGTTCCAGTAACACTGATTCGAATTCAAACAGCGTTCCAGGAAATCCGGCGGTAGCAGCTCTCATGAAGACAGTTCCAAGAGTCGTGATGGAAGTGTATCATCCACTGGCAGTTCCAGTAACACTGATTCAAACTCAAACAGCGCAGGATCCAGTACATCTGGCGGTAGCAGCACTTATGGATACAGCTCCAACAGTCGTGATGGAAGTGTATCATCCACCGGCAGTTCCAGTTACACTGATTCAAACTCAAACAGCGCAGGATCCAGTACATCGGGCGGTAGCAGCACTTATGGATACAGTTCCAACAGTCATGATGGAAGATCCAGGAAATCCGACGGTAGCAGCTCTCATGAAGACAGTTCCAAGAGTCGTGATGAAAGTGTATCGACCACCGGCAGTTCCAGTAACACTGATTCAAACTCAAGCAGCGCAGGATCCAGTACATCCGGTGGTAGCAGCACTTATGGATACAGCTCCAACAGTGGTGATGGAAGTGTATCATCCACCGGCAGTTCCAGTAACACTGAGTCAAACTCAAACAGCGAAGGATCCAGAACATCTGGCGGTAGCAGCACTTATGGATACAGTTCCAACAGTCGTGATGGAAGTGTATCATCCACCGGCAGTTCCAGTAACACTGATTCAAACTCAAACAGCGCAGGATCCAGTACATCTGGTGGTAGCAGCACTTATGGATACAGTTCCAACAGTCGTGATGAAAGTGTATCATCCACCGGCAGTTCCAGTAACACTGATTCGAATTCAAACAGCCGCAGGATCCAGTACATCTGGCAGTAGCAACACTCATGGTGCCAGCCATGGTTTGGCATCATCTGGCGGTAGTATAGCATCAGCAACTAGTACTCCTGACATACTTACCATAGCACTAAGTGAAGACTCTTCCGAGGTGGATATTGATCTTGGCAATTTAGGCTGGTGGTGGAATTCAGACAATAAGGCACAAAGAGCGGCAGGCGGCGCAACAAAGTCTGAAGCTTCATCATCCACTCAAGCTACTACAGAACCGTTTCGTCCACTGGCAGTACCAGTAACACCGATTCAAGCTCAAAAAGTGCAGGATCCCGTACATCCGGCGGTAGCAGCACTTATGGATATAGCTCCAGCCATCGTGGTGGAAGCGTATCATCCACCGGCAGTTCCAGCAACACTGATTCAAGCACAAAGAATGCAGGATCCAGTACATCCGGCGGTAGCAGCACTTATGGATATAGCTCCAGCCATCGTGGTGGAAGCGTATCATCCACCGGCAGTTCCAGCAACACTGATTCAAGCACAAAGAATGCAGGATCCAGTACATCTGGCGGTAGCAGCACTTATGGATATAGCTCTAGCCATCGTGGTGGAAGTGTATCATCCACCGGCAGTTCCAGCAACACTGATTCAAGCACAAAGAGTGCAGGATCCAGTACATCCGGCGGTAGCAGCACTTACGGATATAGCTCCAGGCATCGTGGATCTAGTACATCCGGCGGTAGCAGCACTTATGGATACAGTTCCAACAGTCGTGATGGAAGTGTATCATCCACCGGCAGTTCCAGTAACACTGATTCAAACTCAAACAGCGCGGGATCCAGTACATCCGGTGGTAGCAGCACTTATGGATACAGTTCCAACAGTCGTGATGGAAGTGTATCATCCACCGGCAGTTCCAGTAACACTGATTCAAACTCAAACAGCGCGGGATCCAGTACATCCGGTGGTAGCAGCACTTATGGATACAGTTCCAACAGTCGTGATGGAAGTGTATCATCCACCGGCAGTTCCAGTAACACTGATGCAAGCACAGACCTTACAGGATCCAGTACATCCGGCGGTAGCAGCACTTATGGATACAGTTCCGACAGTCGTGATGGAAGTGTATCATCCACCGGCAGTTCCAGTAACACTGATGCAAGCACAGACCTGGCAGGATCCAGTACATCTGGCGGTAGCAGCACTTATGGATACAGTTCCGACTGTGGTGATGGAAGTGTATCATCCACCGGCAGTTCCAGTAACACTGATGCAAGCACAGACCTTGCAGGATCCAGTACATCCGGCGGTAGCAGCACTTATGGATACAGTTCCGACAGTCGTGATGGAAGTGTATCATCCACCGGCAGTTCCAGTAACACTGATGCAAGCACAGACCTTGCAGGATCCAGTACATCGGGCGGTAGCAGCACTTATGGATACAGTTCCAACAGTCGTGATGGAAGTGTATCATCCACCGGCAGTTCCAGTAACACTGATGCAAGCACAGACCTTACAGGATCCAGTACATCCGGCGGTAGCAGCACTTATGGATATAGCTCAAGCAATCGTGATGGAAGTGTATTGGCCACTGGCAGTTCCAGTAACACTGATGCAAGCACCACAGAAGAATCCACCACGTCCGCTGGTAGCAGCACTGAAGGATATAGTTCCAGTAGCCATGATGGAAGCGTAA
Protein
MRFVLCCTLIALAALSVKAFGHHPGNRDTVEVKNRKYNAASSESSYLNKDNDSISAGAHRAKSVEQSQDKSKYTSGPEGVSYSGRSQNYKDSKQAYADYHSDPNGGSASAGQSRDSSLRERNVHYVSDGEAVAASSDARDENRSAQQNAQANWNADGSYGVSADRSGSASSRRRQANYYSDKDITAASKDDSRADSSRRSNAYYNRDSDGSESAGLSDRRASSSKNDNVFVYRTKDSIGGQAKSSRSSHSQESDAYYNSSPDGSYNAGTRDSSISNKKKASSTIYADKDQIRAANDRYSSKQLKQSSAQISSGPEGTSVSSKDRQYSNDKRSKSDAYVGRDGTVAYSNKDSEKTSRQSNTNYADQNSVRSDSAASDQTSNSYDKGYSDKNTVAHSSGSRGSQNQKSSSYRADKDGFSSSTNTEKSRFSSSNSVVETSDGTSASRESSAEDTKSSNSNVQSDETGEEEELFDVVSYQKIEDGKPVIIMKVIPVEKSASQSSSSRSSQESASYSSSSSSSTEESSSSSSRAASSTDASSNTDSNSNSAGSSTSGGSSTYGYSSNSRDGSVSTTGSSSNTDSNSNSVPGNPAVAALMKTVPRVVMEVYHPLAVPVTLIQTQTAQDPVHLAVAALMDTAPTVVMEVYHPPAVPVTLIQTQTAQDPVHRAVAALMDTVPTVMMEDPGNPTVAALMKTVPRVVMKVYRPPAVPVTLIQTQAAQDPVHPVVAALMDTAPTVVMEVYHPPAVPVTLSQTQTAKDPEHLAVAALMDTVPTVVMEVYHPPAVPVTLIQTQTAQDPVHLVVAALMDTVPTVVMKVYHPPAVPVTLIRIQTAAGSSTSGSSNTHGASHGLASSGGSIASATSTPDILTIALSEDSSEVDIDLGNLGWWWNSDNKAQRAAGGATKSEASSSTQATTEPFRPLAVPVTPIQAQKVQDPVHPAVAALMDIAPAIVVEAYHPPAVPATLIQAQRMQDPVHPAVAALMDIAPAIVVEAYHPPAVPATLIQAQRMQDPVHLAVAALMDIALAIVVEVYHPPAVPATLIQAQRVQDPVHPAVAALTDIAPGIVDLVHPAVAALMDTVPTVVMEVYHPPAVPVTLIQTQTARDPVHPVVAALMDTVPTVVMEVYHPPAVPVTLIQTQTARDPVHPVVAALMDTVPTVVMEVYHPPAVPVTLMQAQTLQDPVHPAVAALMDTVPTVVMEVYHPPAVPVTLMQAQTWQDPVHLAVAALMDTVPTVVMEVYHPPAVPVTLMQAQTLQDPVHPAVAALMDTVPTVVMEVYHPPAVPVTLMQAQTLQDPVHRAVAALMDTVPTVVMEVYHPPAVPVTLMQAQTLQDPVHPAVAALMDIAQAIVMEVYWPLAVPVTLMQAPQKNPPRPLVAALKDIVPVAMMEA
Summary
Uniprot
ProteinModelPortal
PDB
2AAA
E-value=0.0945448,
Score=88
Ontologies
GO
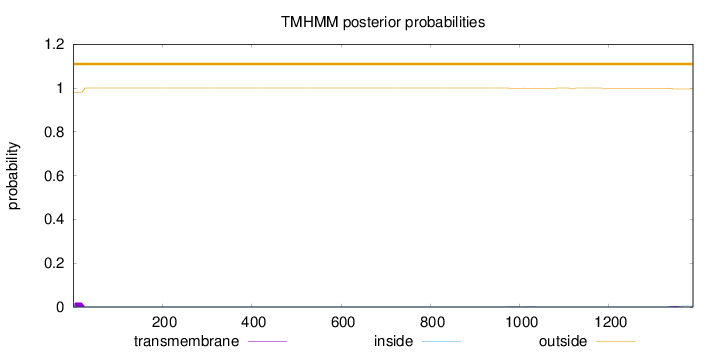
Topology
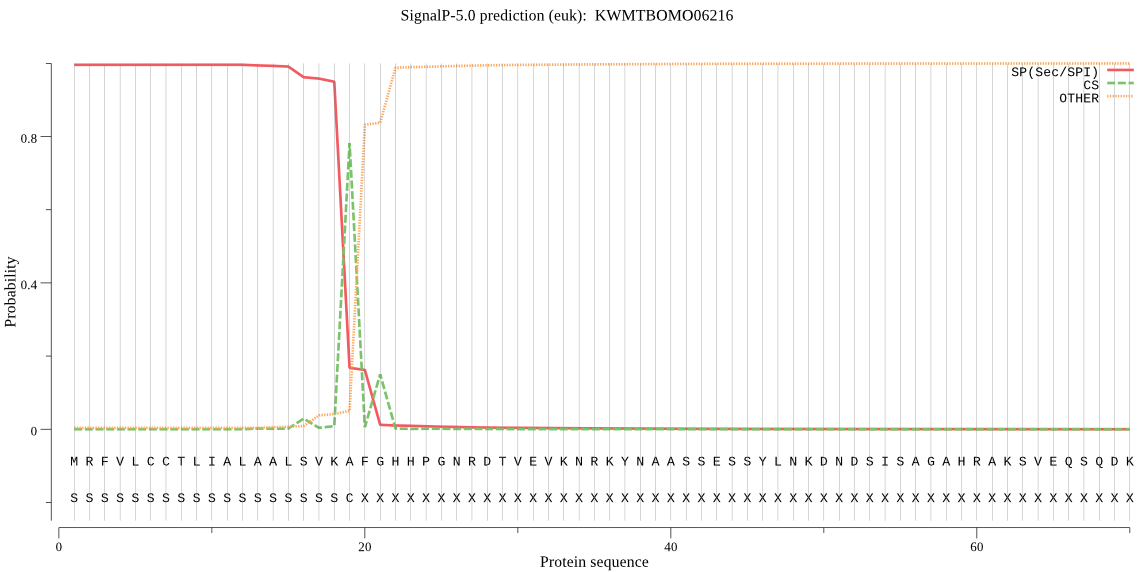
SignalP
Position: 1 - 19,
Likelihood: 0.995821
Length:
1388
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.532870000000001
Exp number, first 60 AAs:
0.41654
Total prob of N-in:
0.02046
outside
1 - 1388
Population Genetic Test Statistics
Pi
25.484731
Theta
203.386352
Tajima's D
0.590859
CLR
0.540146
CSRT
0.534073296335183
Interpretation
Uncertain
