Gene
KWMTBOMO06211
Pre Gene Modal
BGIBMGA001792
Annotation
PREDICTED:_protein_artemis-like_isoform_X2_[Bombyx_mori]
Full name
Protein artemis
Alternative Name
DNA cross-link repair 1C protein
SNM1-like protein
Protein A-SCID
SNM1 homolog C
SNM1-like protein
Protein A-SCID
SNM1 homolog C
Location in the cell
Cytoplasmic Reliability : 1.137 Nuclear Reliability : 1.327
Sequence
CDS
ATGTCCGAGGTCTCGGCGACTATCGTGGAGTATGAAAGCAATTGTGGACCCATCATGAGACACATAAGAACATTAAAGTTGGAATCAACAACAATAACTTTAATAATGGATGACGGGAGCCATAAGTATTTGAATGTCAAAACGATACCGGCCGGCCACTGCCTCGGATCTGTCATGTTTCTATTTGAGATAAACAATCAGACGATACTGTACACAGGGGACTTCAGAATGAATCCAGAGAACATATCTGCTTTTGGACAACTGCACAAAGATAACATGCCAATTAAAATAAACACAATTTACCTTGATACCACATTCCAGAATGAAAGTTTTGACAATTTCCCACGCAGGAAAGACAGCATCCGAATGTTGGTCAATCATATAAAGCAGTGGGTAGACGGAGAACCTACCAACAGAATAGCCTTGCATACATCTGCTCGTTATGGCTACGAATTTGTTTTCAATGAAATCTACAACATTTTAAACATGAAGACTTATGTTAGTGATGATAAATGGGCATTGTACAGCAAATTACCATCTGTTAAAGGGATCACGAACCACAGCGAAGATACGATGATACATTTATCAATGAGACACCGTTCAATGGACAATGAAAACTCGCCCTTAGACAGCCGAGACGGCGTTATATATGTATGTTTCGCGACGCATTGCAGCCGAGAGGAATTGAATTTTTTCGTCAGTTATTTTACACCGGACAAGGTTGTCGGTTTCAGGGATCAGTTTATACCGAAAGCGGGCACGAAACGATGTCACGTCTTTGAGCCAACGAGAACGGTGAAGATTCGTAAATTCAATAACAATAATGCATTTAATAAAATCGATCCGAGCATTGTAAATGATTTATTTTGA
Protein
MSEVSATIVEYESNCGPIMRHIRTLKLESTTITLIMDDGSHKYLNVKTIPAGHCLGSVMFLFEINNQTILYTGDFRMNPENISAFGQLHKDNMPIKINTIYLDTTFQNESFDNFPRRKDSIRMLVNHIKQWVDGEPTNRIALHTSARYGYEFVFNEIYNILNMKTYVSDDKWALYSKLPSVKGITNHSEDTMIHLSMRHRSMDNENSPLDSRDGVIYVCFATHCSREELNFFVSYFTPDKVVGFRDQFIPKAGTKRCHVFEPTRTVKIRKFNNNNAFNKIDPSIVNDLF
Summary
Description
Required for V(D)J recombination, the process by which exons encoding the antigen-binding domains of immunoglobulins and T-cell receptor proteins are assembled from individual V, (D), and J gene segments. V(D)J recombination is initiated by the lymphoid specific RAG endonuclease complex, which generates site specific DNA double strand breaks (DSBs). These DSBs present two types of DNA end structures: hairpin sealed coding ends and phosphorylated blunt signal ends. These ends are independently repaired by the non homologous end joining (NHEJ) pathway to form coding and signal joints respectively. This protein likely exhibits single-strand specific 5'-3' exonuclease activity in isolation, and may acquire endonucleolytic activity on 5' and 3' hairpins and overhangs when in a complex with PRKDC. The latter activity may be required specifically for the resolution of closed hairpins prior to the formation of the coding joint. May also be required for the repair of complex DSBs induced by ionizing radiation, which require substantial end-processing prior to religation by NHEJ.
Required for V(D)J recombination, the process by which exons encoding the antigen-binding domains of immunoglobulins and T-cell receptor proteins are assembled from individual V, (D), and J gene segments. V(D)J recombination is initiated by the lymphoid specific RAG endonuclease complex, which generates site specific DNA double strand breaks (DSBs). These DSBs present two types of DNA end structures: hairpin sealed coding ends and phosphorylated blunt signal ends. These ends are independently repaired by the non homologous end joining (NHEJ) pathway to form coding and signal joints respectively. This protein exhibits single-strand specific 5'-3' exonuclease activity in isolation and acquires endonucleolytic activity on 5' and 3' hairpins and overhangs when in a complex with PRKDC. The latter activity is required specifically for the resolution of closed hairpins prior to the formation of the coding joint. May also be required for the repair of complex DSBs induced by ionizing radiation, which require substantial end-processing prior to religation by NHEJ.
Required for V(D)J recombination, the process by which exons encoding the antigen-binding domains of immunoglobulins and T-cell receptor proteins are assembled from individual V, (D), and J gene segments. V(D)J recombination is initiated by the lymphoid specific RAG endonuclease complex, which generates site specific DNA double strand breaks (DSBs). These DSBs present two types of DNA end structures: hairpin sealed coding ends and phosphorylated blunt signal ends. These ends are independently repaired by the non homologous end joining (NHEJ) pathway to form coding and signal joints respectively. This protein exhibits single-strand specific 5'-3' exonuclease activity in isolation and acquires endonucleolytic activity on 5' and 3' hairpins and overhangs when in a complex with PRKDC. The latter activity is required specifically for the resolution of closed hairpins prior to the formation of the coding joint. May also be required for the repair of complex DSBs induced by ionizing radiation, which require substantial end-processing prior to religation by NHEJ.
Subunit
Interacts with ATM, BRCA1, PRKDC and TP53BP1. Also exhibits ATM- and phosphorylation-dependent interaction with the MRN complex, composed of MRE11, RAD50, and NBN (By similarity).
Interacts with ATM, BRCA1, PRKDC and TP53BP1. Also exhibits ATM- and phosphorylation-dependent interaction with the MRN complex, composed of MRE11, RAD50, and NBN.
Interacts with ATM, BRCA1, PRKDC and TP53BP1. Also exhibits ATM- and phosphorylation-dependent interaction with the MRN complex, composed of MRE11, RAD50, and NBN.
Similarity
Belongs to the EF-1-beta/EF-1-delta family.
Belongs to the DNA repair metallo-beta-lactamase (DRMBL) family.
Belongs to the DNA repair metallo-beta-lactamase (DRMBL) family.
Keywords
Adaptive immunity
Alternative splicing
Complete proteome
DNA damage
DNA recombination
DNA repair
Endonuclease
Exonuclease
Hydrolase
Immunity
Magnesium
Nuclease
Nucleus
Phosphoprotein
Reference proteome
SCID
3D-structure
Disease mutation
Polymorphism
Feature
chain Protein artemis
splice variant In isoform 3.
sequence variant In Omenn syndrome; dbSNP:rs121908159.
splice variant In isoform 3.
sequence variant In Omenn syndrome; dbSNP:rs121908159.
Uniprot
A0A194QAR3
A0A2W1BHH9
A0A1E1WS68
A0A212EZM6
A0A2H1WF70
N6UC24
+ More
K7JPN7 A0A1S3DLK9 A0A2Z5TRC6 A0A067R1R8 A0A087ZVH9 A0A2J7PUM3 D6WZE5 A0A2A3ER42 A0A151I7T0 A0A1Y1N5K8 A0A195DZG0 A0A2P8ZJQ1 A0A0M8ZZG6 F4WCY0 A0A310SFB5 A0A232F3W7 A0A1W4WSD0 E2C6B8 B0X3C4 A0A154PP67 A0A212CC14 A0A151JY47 A0A1B0DC75 A0A158NB18 A0A1B6E9Y2 A0A1B6J9C3 A0A1D2MW33 A0A151WXZ2 V4A3J4 C3YPJ3 Q32MX8 S4RFM2 A0A1D1V1R7 A0A0T6B8H2 Q8K4J0-2 A0A2J8QJP2 A0A2I2ZB06 A0A2J8VKL4 Q96SD1-4 A0A2K6AKS5 A0A2K6ND80 A0A182N057 E5FGH7 E5FGH6 A0A2K6JQ38 A0A2K6JQ50 E5FGI2 E5FGH9 E5FGI3 A0A2K5X8W2 A0A1D5QCS2 E5FGH5 A0A2K5X8T2 E5FGG9
K7JPN7 A0A1S3DLK9 A0A2Z5TRC6 A0A067R1R8 A0A087ZVH9 A0A2J7PUM3 D6WZE5 A0A2A3ER42 A0A151I7T0 A0A1Y1N5K8 A0A195DZG0 A0A2P8ZJQ1 A0A0M8ZZG6 F4WCY0 A0A310SFB5 A0A232F3W7 A0A1W4WSD0 E2C6B8 B0X3C4 A0A154PP67 A0A212CC14 A0A151JY47 A0A1B0DC75 A0A158NB18 A0A1B6E9Y2 A0A1B6J9C3 A0A1D2MW33 A0A151WXZ2 V4A3J4 C3YPJ3 Q32MX8 S4RFM2 A0A1D1V1R7 A0A0T6B8H2 Q8K4J0-2 A0A2J8QJP2 A0A2I2ZB06 A0A2J8VKL4 Q96SD1-4 A0A2K6AKS5 A0A2K6ND80 A0A182N057 E5FGH7 E5FGH6 A0A2K6JQ38 A0A2K6JQ50 E5FGI2 E5FGH9 E5FGI3 A0A2K5X8W2 A0A1D5QCS2 E5FGH5 A0A2K5X8T2 E5FGG9
EC Number
3.1.-.-
Pubmed
26354079
28756777
22118469
23537049
20075255
26760975
+ More
24845553 18362917 19820115 28004739 29403074 21719571 28648823 20798317 21347285 27289101 23254933 18563158 15489334 19468303 27649274 15699179 16141072 12504013 12177301 12615897 21183079 22398555 11336668 12055248 14702039 15164054 11955432 15071507 15468306 14744996 15574326 15574327 15456891 15723659 15811628 15936993 19690332 23186163 12406895 12921762 12592555 12569164 15731174 25362486 20975951 17431167
24845553 18362917 19820115 28004739 29403074 21719571 28648823 20798317 21347285 27289101 23254933 18563158 15489334 19468303 27649274 15699179 16141072 12504013 12177301 12615897 21183079 22398555 11336668 12055248 14702039 15164054 11955432 15071507 15468306 14744996 15574326 15574327 15456891 15723659 15811628 15936993 19690332 23186163 12406895 12921762 12592555 12569164 15731174 25362486 20975951 17431167
EMBL
KQ459463
KPJ00511.1
KZ150374
PZC71123.1
GDQN01001185
JAT89869.1
+ More
AGBW02011242 OWR46940.1 ODYU01008272 SOQ51713.1 APGK01029568 KB740735 KB632302 ENN79205.1 ERL91777.1 FX985783 BBA93670.1 KK852984 KDR12848.1 NEVH01021195 PNF20023.1 KQ971372 EFA09718.1 KZ288193 PBC34160.1 KQ978396 KYM94225.1 GEZM01015454 GEZM01015453 JAV91636.1 KQ980050 KYN18014.1 PYGN01000035 PSN56700.1 KQ435792 KOX74265.1 GL888074 EGI67961.1 KQ765174 OAD54108.1 NNAY01001022 OXU25454.1 GL453058 EFN76517.1 DS232312 EDS39816.1 KQ435007 KZC13681.1 MKHE01000023 OWK03384.1 KQ981533 KYN40113.1 AJVK01030620 ADTU01010746 GEDC01002593 JAS34705.1 GECU01037315 GECU01027925 GECU01016805 GECU01011934 JAS70391.1 JAS79781.1 JAS90901.1 JAS95772.1 LJIJ01000479 ODM97014.1 KQ982665 KYQ52561.1 KB202283 ESO91307.1 GG666538 EEN57777.1 AL732620 BC108935 AAI08936.1 BDGG01000002 GAU93527.1 LJIG01009162 KRT83632.1 AF387731 AK037126 AK052369 AK088810 NBAG03000034 PNI96437.1 PNI96439.1 CABD030068136 CABD030068137 CABD030068138 CABD030068139 CABD030068140 NDHI03003418 PNJ58068.1 AJ296101 AF395747 AF395748 AF395749 AF395750 AF395751 AF395752 AK021422 DQ504427 AC069544 AL360083 CH471072 BC000863 BC009185 BC022254 HM486784 ADQ01120.1 HM486783 ADQ01119.1 HM486789 ADQ01125.1 HM486786 ADQ01122.1 AHZZ02034837 AHZZ02034838 HM486790 ADQ01126.1 AQIA01070118 AQIA01070119 AQIA01070120 AQIA01070121 AQIA01070122 AQIA01070123 JSUE03042906 JSUE03042907 JSUE03042908 HM486782 ADQ01118.1 HM486776 ADQ01112.1
AGBW02011242 OWR46940.1 ODYU01008272 SOQ51713.1 APGK01029568 KB740735 KB632302 ENN79205.1 ERL91777.1 FX985783 BBA93670.1 KK852984 KDR12848.1 NEVH01021195 PNF20023.1 KQ971372 EFA09718.1 KZ288193 PBC34160.1 KQ978396 KYM94225.1 GEZM01015454 GEZM01015453 JAV91636.1 KQ980050 KYN18014.1 PYGN01000035 PSN56700.1 KQ435792 KOX74265.1 GL888074 EGI67961.1 KQ765174 OAD54108.1 NNAY01001022 OXU25454.1 GL453058 EFN76517.1 DS232312 EDS39816.1 KQ435007 KZC13681.1 MKHE01000023 OWK03384.1 KQ981533 KYN40113.1 AJVK01030620 ADTU01010746 GEDC01002593 JAS34705.1 GECU01037315 GECU01027925 GECU01016805 GECU01011934 JAS70391.1 JAS79781.1 JAS90901.1 JAS95772.1 LJIJ01000479 ODM97014.1 KQ982665 KYQ52561.1 KB202283 ESO91307.1 GG666538 EEN57777.1 AL732620 BC108935 AAI08936.1 BDGG01000002 GAU93527.1 LJIG01009162 KRT83632.1 AF387731 AK037126 AK052369 AK088810 NBAG03000034 PNI96437.1 PNI96439.1 CABD030068136 CABD030068137 CABD030068138 CABD030068139 CABD030068140 NDHI03003418 PNJ58068.1 AJ296101 AF395747 AF395748 AF395749 AF395750 AF395751 AF395752 AK021422 DQ504427 AC069544 AL360083 CH471072 BC000863 BC009185 BC022254 HM486784 ADQ01120.1 HM486783 ADQ01119.1 HM486789 ADQ01125.1 HM486786 ADQ01122.1 AHZZ02034837 AHZZ02034838 HM486790 ADQ01126.1 AQIA01070118 AQIA01070119 AQIA01070120 AQIA01070121 AQIA01070122 AQIA01070123 JSUE03042906 JSUE03042907 JSUE03042908 HM486782 ADQ01118.1 HM486776 ADQ01112.1
Proteomes
UP000053268
UP000007151
UP000019118
UP000030742
UP000002358
UP000079169
+ More
UP000027135 UP000005203 UP000235965 UP000007266 UP000242457 UP000078542 UP000078492 UP000245037 UP000053105 UP000007755 UP000215335 UP000192223 UP000008237 UP000002320 UP000076502 UP000078541 UP000092462 UP000005205 UP000094527 UP000075809 UP000030746 UP000001554 UP000000589 UP000245300 UP000186922 UP000001519 UP000005640 UP000233140 UP000233200 UP000075884 UP000233180 UP000028761 UP000233100 UP000006718
UP000027135 UP000005203 UP000235965 UP000007266 UP000242457 UP000078542 UP000078492 UP000245037 UP000053105 UP000007755 UP000215335 UP000192223 UP000008237 UP000002320 UP000076502 UP000078541 UP000092462 UP000005205 UP000094527 UP000075809 UP000030746 UP000001554 UP000000589 UP000245300 UP000186922 UP000001519 UP000005640 UP000233140 UP000233200 UP000075884 UP000233180 UP000028761 UP000233100 UP000006718
Pfam
Interpro
IPR011084
DRMBL
+ More
IPR036866 RibonucZ/Hydroxyglut_hydro
IPR038579 Ribosomal_S21e_sf
IPR001931 Ribosomal_S21e
IPR018279 Ribosomal_S21e_CS
IPR001279 Metallo-B-lactamas
IPR014038 EF1B_bsu/dsu_GNE
IPR018940 EF-1_beta_acid_region_euk
IPR001326 Transl_elong_EF1B_B/D_CS
IPR014717 Transl_elong_EF1B/ribosomal_S6
IPR036219 eEF-1beta-like_sf
IPR036866 RibonucZ/Hydroxyglut_hydro
IPR038579 Ribosomal_S21e_sf
IPR001931 Ribosomal_S21e
IPR018279 Ribosomal_S21e_CS
IPR001279 Metallo-B-lactamas
IPR014038 EF1B_bsu/dsu_GNE
IPR018940 EF-1_beta_acid_region_euk
IPR001326 Transl_elong_EF1B_B/D_CS
IPR014717 Transl_elong_EF1B/ribosomal_S6
IPR036219 eEF-1beta-like_sf
Gene 3D
CDD
ProteinModelPortal
A0A194QAR3
A0A2W1BHH9
A0A1E1WS68
A0A212EZM6
A0A2H1WF70
N6UC24
+ More
K7JPN7 A0A1S3DLK9 A0A2Z5TRC6 A0A067R1R8 A0A087ZVH9 A0A2J7PUM3 D6WZE5 A0A2A3ER42 A0A151I7T0 A0A1Y1N5K8 A0A195DZG0 A0A2P8ZJQ1 A0A0M8ZZG6 F4WCY0 A0A310SFB5 A0A232F3W7 A0A1W4WSD0 E2C6B8 B0X3C4 A0A154PP67 A0A212CC14 A0A151JY47 A0A1B0DC75 A0A158NB18 A0A1B6E9Y2 A0A1B6J9C3 A0A1D2MW33 A0A151WXZ2 V4A3J4 C3YPJ3 Q32MX8 S4RFM2 A0A1D1V1R7 A0A0T6B8H2 Q8K4J0-2 A0A2J8QJP2 A0A2I2ZB06 A0A2J8VKL4 Q96SD1-4 A0A2K6AKS5 A0A2K6ND80 A0A182N057 E5FGH7 E5FGH6 A0A2K6JQ38 A0A2K6JQ50 E5FGI2 E5FGH9 E5FGI3 A0A2K5X8W2 A0A1D5QCS2 E5FGH5 A0A2K5X8T2 E5FGG9
K7JPN7 A0A1S3DLK9 A0A2Z5TRC6 A0A067R1R8 A0A087ZVH9 A0A2J7PUM3 D6WZE5 A0A2A3ER42 A0A151I7T0 A0A1Y1N5K8 A0A195DZG0 A0A2P8ZJQ1 A0A0M8ZZG6 F4WCY0 A0A310SFB5 A0A232F3W7 A0A1W4WSD0 E2C6B8 B0X3C4 A0A154PP67 A0A212CC14 A0A151JY47 A0A1B0DC75 A0A158NB18 A0A1B6E9Y2 A0A1B6J9C3 A0A1D2MW33 A0A151WXZ2 V4A3J4 C3YPJ3 Q32MX8 S4RFM2 A0A1D1V1R7 A0A0T6B8H2 Q8K4J0-2 A0A2J8QJP2 A0A2I2ZB06 A0A2J8VKL4 Q96SD1-4 A0A2K6AKS5 A0A2K6ND80 A0A182N057 E5FGH7 E5FGH6 A0A2K6JQ38 A0A2K6JQ50 E5FGI2 E5FGH9 E5FGI3 A0A2K5X8W2 A0A1D5QCS2 E5FGH5 A0A2K5X8T2 E5FGG9
PDB
5AHO
E-value=7.26281e-10,
Score=151
Ontologies
GO
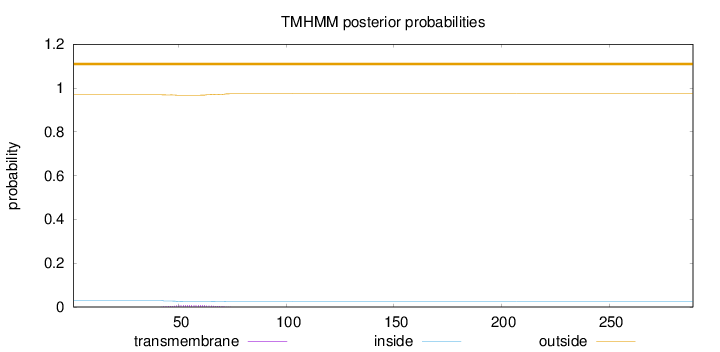
Topology
Subcellular location
Nucleus
Length:
289
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18884
Exp number, first 60 AAs:
0.12707
Total prob of N-in:
0.03076
outside
1 - 289
Population Genetic Test Statistics
Pi
197.374724
Theta
148.098268
Tajima's D
0.95159
CLR
17.402698
CSRT
0.646267686615669
Interpretation
Possibly Positive selection
