Gene
KWMTBOMO06199
Pre Gene Modal
BGIBMGA001780
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.936
Sequence
CDS
ATGTCCAACGGTATGAAGGATTATTATCACAGAGTCGTATCTAATGAGGGAGATGATGATGATGATGATGAGGGAAACAAGTACAGACCGCTGTTGAGGCAGATCCTCATAACGAGCCCCGCCTGGATGTTGCTGTTCGCTACAGGCTTTTCATTCGGCGCTCCGACCGTGTTCATACCTCAAATTCGGAAGGAAAAGAATTCGACCGAAGCGATCAGTGACGAAATGGCTTCGTGGCTATCTTCCGTGTTCGGATATAGCTCTCTGCCGTGGATCATCGTTATCTGCTACGCCATTACGCGCTTAGGAAGAAAGATTCCTTTCATGTTCGTCTCCATAAACACGTTCGTCTTCAACGTCATCTTATATTTCAGCACTAGCAACACCCAAATGTTAATTAGCGAGATACTACAGGGCATGAATCACGGCGCCAACTTAACTTTGACTATAATAATACTGTCCGAATACACTTCGCCGAAATACAGAGGTGTCTTCTTAACAATAAAATCCGCCACCTTCTATTGGGGTGTGTGGTGCGCGAACGCAATTGGAACATTCCTCCATTGGAAATATATTCCGTTGACAGCCATCGTGTTGATATTATACAATTTCGTCTCGTTTTTGTGGCCGGAGTCGCCTCACTGGTTGGCCAGCGAAGGTAGGTTCGAGGAATGCGCCCGATCATACTTCTGGTTGATGGGTAAAAACGAGTCAACGATACAAGAATTGGAGAGACTCGTTAATTCGCAAAAAGAATGCATTAAAAATCGTAGGACGTTCGATAGGCACAGAATATTCGAGATCTTGACCGCGAAGCTGTTTTACAAACCGACATTAGTGTGTATAGTTATGATGCTGCAATATAATTTCACCGGTAAAATCGTGTGTGCAATGTACGCCATTGATATCATAAAACGCATAACTGGAAGCGAATCGACCGCGTATTACGTGATGCTGCTACTAGATGGGGTTACTGTATTGGGCATGTACTTCGGTTGTTTTTTAACGAGAGTGGTCAAGCGACGCTCTTTACTAATGACCTCGGGCACGACAAGTGTAATATTTTTATGCATCCTATCCTTGTATTTGTATTTAATAAATTTAGCGTTTATAAACGAATCTAGATTTGTAACTATAGGTTTGTTAGTCGGATTCTCGTTGAGCGTTAGCATTGGACCGGTGATTTTGTTAATATCAACGCACGCTGAGCTGACGCCATTAAAATACAAAAGTCATTGTATGGCCGTTGCCGCGATTTCATACAATATCGTCTCCGGTACATTATTGAAAATGTCGCCGTATATCTTTTTGGCTTTCGGGATGCACGGTACGTTTTTATTTTACGGTCTCACTTCGGGATCCTGTCTATTGATACTTTACTTCTGTTTACCGGAAACAAAAGACAAAACCCTGCAGGAAATCGAAGATTATTTCGAAGGTGAAGAAAAAAAAACGTGTAATTCAAGTTTAATTTAA
Protein
MSNGMKDYYHRVVSNEGDDDDDDEGNKYRPLLRQILITSPAWMLLFATGFSFGAPTVFIPQIRKEKNSTEAISDEMASWLSSVFGYSSLPWIIVICYAITRLGRKIPFMFVSINTFVFNVILYFSTSNTQMLISEILQGMNHGANLTLTIIILSEYTSPKYRGVFLTIKSATFYWGVWCANAIGTFLHWKYIPLTAIVLILYNFVSFLWPESPHWLASEGRFEECARSYFWLMGKNESTIQELERLVNSQKECIKNRRTFDRHRIFEILTAKLFYKPTLVCIVMMLQYNFTGKIVCAMYAIDIIKRITGSESTAYYVMLLLDGVTVLGMYFGCFLTRVVKRRSLLMTSGTTSVIFLCILSLYLYLINLAFINESRFVTIGLLVGFSLSVSIGPVILLISTHAELTPLKYKSHCMAVAAISYNIVSGTLLKMSPYIFLAFGMHGTFLFYGLTSGSCLLILYFCLPETKDKTLQEIEDYFEGEEKKTCNSSLI
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9IWZ7
A0A0L7KPS0
A0A2W1BJN4
H9JAI4
A0A0L7KW08
A0A2H1VQR0
+ More
A0A2A4JBS9 H9JFD5 A0A2W1BR72 A0A2H1X3V1 A0A2H1X3Y1 A0A2W1BJK8 A0A2W1BLU7 A0A2W1BQ15 A0A2H1VQR9 A0A2H1VYW0 A0A2A4K454 A0A2A4K3Q9 A0A2H1VYX5 A0A0L7LBU2 A0A0L7KRG0 A0A0L7LW97 A0A0L7LSQ8 A0A194Q2H3 A0A212EJQ6 A0A194R9C4 A0A0N1IQT4 A0A212ETB8 A0A2H1WKZ9 A0A2A4KA42 A0A2W1B9E8 A0A2A4KB29 A0A2H1WK46 A0A2W1BN20 A0A2W1BF54 A0A2H1WX63 A0A2A4JKJ9 A0A194QKY0 H9J017 A0A194QJ23 A0A2A4JJR5 A0A0L7LC11 A0A0L7LHM6 A0A194QJ80 A0A0L7KWB8 A0A2A4JT91 A0A0N1IG19 A0A212F9C9 A0A0N1IPS2 A0A0L7LCK3 H9J4X7 A0A0N1I9K7 A0A2A4IVM9 A0A194QJI7 A0A194QHU0 A0A023PNK9 A0A0N0PAH4 A0A194R9K8 A0A2A4JTG8 A0A2H1WVP2 A0A2W1BFI0 A0A2H1WB04 A0A2A4J748 B0W5I2 A0A2A4JNG3 A0A0N0PEP7 A0A0L7L7N1 A0A2H1WAU6 A0A194QEH9 A0A0L7KLX5 A0A212ENJ2 A0A0L7LIB6 A0A212F9B1 A0A194QPY2 A0A0N0PEE0 A0A212F161 A0A0N1IHN6 A0A2W1BMU2 A0A3S2LEF2 A0A336MDU0 A0A0N0PE20 A0A212FJ74 H9J525 H9J4X3 A0A194QJ30 A0A182GCQ0 A0A182GTB3 H9IV67 A0A2A4JUU3 A0A2W1BD78 H9J4X0 A0A0N1IDG3 A0A2H1W936 A0A194QJ40 Q17LS5
A0A2A4JBS9 H9JFD5 A0A2W1BR72 A0A2H1X3V1 A0A2H1X3Y1 A0A2W1BJK8 A0A2W1BLU7 A0A2W1BQ15 A0A2H1VQR9 A0A2H1VYW0 A0A2A4K454 A0A2A4K3Q9 A0A2H1VYX5 A0A0L7LBU2 A0A0L7KRG0 A0A0L7LW97 A0A0L7LSQ8 A0A194Q2H3 A0A212EJQ6 A0A194R9C4 A0A0N1IQT4 A0A212ETB8 A0A2H1WKZ9 A0A2A4KA42 A0A2W1B9E8 A0A2A4KB29 A0A2H1WK46 A0A2W1BN20 A0A2W1BF54 A0A2H1WX63 A0A2A4JKJ9 A0A194QKY0 H9J017 A0A194QJ23 A0A2A4JJR5 A0A0L7LC11 A0A0L7LHM6 A0A194QJ80 A0A0L7KWB8 A0A2A4JT91 A0A0N1IG19 A0A212F9C9 A0A0N1IPS2 A0A0L7LCK3 H9J4X7 A0A0N1I9K7 A0A2A4IVM9 A0A194QJI7 A0A194QHU0 A0A023PNK9 A0A0N0PAH4 A0A194R9K8 A0A2A4JTG8 A0A2H1WVP2 A0A2W1BFI0 A0A2H1WB04 A0A2A4J748 B0W5I2 A0A2A4JNG3 A0A0N0PEP7 A0A0L7L7N1 A0A2H1WAU6 A0A194QEH9 A0A0L7KLX5 A0A212ENJ2 A0A0L7LIB6 A0A212F9B1 A0A194QPY2 A0A0N0PEE0 A0A212F161 A0A0N1IHN6 A0A2W1BMU2 A0A3S2LEF2 A0A336MDU0 A0A0N0PE20 A0A212FJ74 H9J525 H9J4X3 A0A194QJ30 A0A182GCQ0 A0A182GTB3 H9IV67 A0A2A4JUU3 A0A2W1BD78 H9J4X0 A0A0N1IDG3 A0A2H1W936 A0A194QJ40 Q17LS5
EMBL
BABH01021167
JTDY01007576
KOB65105.1
KZ150004
PZC75259.1
BABH01009421
+ More
JTDY01005057 KOB67418.1 ODYU01003864 SOQ43159.1 NWSH01001973 PCG69525.1 BABH01038306 PZC75260.1 ODYU01013263 SOQ59969.1 SOQ59968.1 PZC75262.1 PZC75261.1 KZ149985 PZC75704.1 SOQ43160.1 ODYU01005293 SOQ46008.1 NWSH01000180 PCG78688.1 PCG78689.1 ODYU01005292 SOQ46007.1 JTDY01001793 KOB72864.1 JTDY01006862 KOB65636.1 JTDY01000013 KOB79431.1 JTDY01000181 KOB78419.1 KQ459589 KPI97605.1 AGBW02014405 OWR41733.1 KQ460500 KPJ14232.1 LADJ01036943 KPJ21300.1 AGBW02012599 OWR44736.1 ODYU01009373 SOQ53753.1 NWSH01000008 PCG80949.1 KZ150249 PZC71838.1 PCG80950.1 ODYU01009098 SOQ53272.1 KZ150067 PZC74120.1 PZC71837.1 ODYU01011726 SOQ57638.1 NWSH01001160 PCG72306.1 KQ458671 KPJ05560.1 BABH01018801 KPJ05558.1 PCG72307.1 JTDY01001770 KOB72935.1 JTDY01001035 KOB75068.1 KPJ05572.1 JTDY01005064 KOB67410.1 NWSH01000704 PCG74683.1 KQ460077 KPJ17891.1 AGBW02009614 OWR50323.1 KPJ17881.1 KOB72931.1 BABH01025717 KPJ17880.1 NWSH01006268 PCG63546.1 KPJ05554.1 KQ458981 KPJ04505.1 KJ439227 KZ150491 AHX25878.1 PZC70688.1 KQ458842 KPJ04897.1 KQ460761 KPJ12541.1 PCG74682.1 ODYU01011356 SOQ57052.1 KZ150133 PZC73021.1 ODYU01007347 SOQ50032.1 NWSH01002771 PCG67556.1 DS231843 EDS35465.1 NWSH01001042 PCG72950.1 KQ459890 KPJ19577.1 JTDY01002470 KOB71349.1 ODYU01007122 SOQ49614.1 KQ459299 KPJ01871.1 JTDY01009224 KOB63914.1 AGBW02013662 OWR43027.1 JTDY01001021 KOB75109.1 OWR50324.1 KPJ05561.1 KPJ19576.1 AGBW02010956 OWR47467.1 KPJ17887.1 KZ150062 PZC74196.1 RSAL01000181 RVE44926.1 UFQT01001016 SSX28504.1 KQ460126 KPJ17588.1 AGBW02008317 OWR53759.1 BABH01038881 BABH01025723 KPJ05563.1 JXUM01009472 KQ560284 KXJ83288.1 JXUM01086634 KQ563580 KXJ73638.1 BABH01000587 NWSH01000578 PCG75536.1 PZC71835.1 BABH01025733 KQ459158 KPJ03590.1 SOQ49615.1 KPJ05573.1 CH477213 EAT47626.1
JTDY01005057 KOB67418.1 ODYU01003864 SOQ43159.1 NWSH01001973 PCG69525.1 BABH01038306 PZC75260.1 ODYU01013263 SOQ59969.1 SOQ59968.1 PZC75262.1 PZC75261.1 KZ149985 PZC75704.1 SOQ43160.1 ODYU01005293 SOQ46008.1 NWSH01000180 PCG78688.1 PCG78689.1 ODYU01005292 SOQ46007.1 JTDY01001793 KOB72864.1 JTDY01006862 KOB65636.1 JTDY01000013 KOB79431.1 JTDY01000181 KOB78419.1 KQ459589 KPI97605.1 AGBW02014405 OWR41733.1 KQ460500 KPJ14232.1 LADJ01036943 KPJ21300.1 AGBW02012599 OWR44736.1 ODYU01009373 SOQ53753.1 NWSH01000008 PCG80949.1 KZ150249 PZC71838.1 PCG80950.1 ODYU01009098 SOQ53272.1 KZ150067 PZC74120.1 PZC71837.1 ODYU01011726 SOQ57638.1 NWSH01001160 PCG72306.1 KQ458671 KPJ05560.1 BABH01018801 KPJ05558.1 PCG72307.1 JTDY01001770 KOB72935.1 JTDY01001035 KOB75068.1 KPJ05572.1 JTDY01005064 KOB67410.1 NWSH01000704 PCG74683.1 KQ460077 KPJ17891.1 AGBW02009614 OWR50323.1 KPJ17881.1 KOB72931.1 BABH01025717 KPJ17880.1 NWSH01006268 PCG63546.1 KPJ05554.1 KQ458981 KPJ04505.1 KJ439227 KZ150491 AHX25878.1 PZC70688.1 KQ458842 KPJ04897.1 KQ460761 KPJ12541.1 PCG74682.1 ODYU01011356 SOQ57052.1 KZ150133 PZC73021.1 ODYU01007347 SOQ50032.1 NWSH01002771 PCG67556.1 DS231843 EDS35465.1 NWSH01001042 PCG72950.1 KQ459890 KPJ19577.1 JTDY01002470 KOB71349.1 ODYU01007122 SOQ49614.1 KQ459299 KPJ01871.1 JTDY01009224 KOB63914.1 AGBW02013662 OWR43027.1 JTDY01001021 KOB75109.1 OWR50324.1 KPJ05561.1 KPJ19576.1 AGBW02010956 OWR47467.1 KPJ17887.1 KZ150062 PZC74196.1 RSAL01000181 RVE44926.1 UFQT01001016 SSX28504.1 KQ460126 KPJ17588.1 AGBW02008317 OWR53759.1 BABH01038881 BABH01025723 KPJ05563.1 JXUM01009472 KQ560284 KXJ83288.1 JXUM01086634 KQ563580 KXJ73638.1 BABH01000587 NWSH01000578 PCG75536.1 PZC71835.1 BABH01025733 KQ459158 KPJ03590.1 SOQ49615.1 KPJ05573.1 CH477213 EAT47626.1
Proteomes
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9IWZ7
A0A0L7KPS0
A0A2W1BJN4
H9JAI4
A0A0L7KW08
A0A2H1VQR0
+ More
A0A2A4JBS9 H9JFD5 A0A2W1BR72 A0A2H1X3V1 A0A2H1X3Y1 A0A2W1BJK8 A0A2W1BLU7 A0A2W1BQ15 A0A2H1VQR9 A0A2H1VYW0 A0A2A4K454 A0A2A4K3Q9 A0A2H1VYX5 A0A0L7LBU2 A0A0L7KRG0 A0A0L7LW97 A0A0L7LSQ8 A0A194Q2H3 A0A212EJQ6 A0A194R9C4 A0A0N1IQT4 A0A212ETB8 A0A2H1WKZ9 A0A2A4KA42 A0A2W1B9E8 A0A2A4KB29 A0A2H1WK46 A0A2W1BN20 A0A2W1BF54 A0A2H1WX63 A0A2A4JKJ9 A0A194QKY0 H9J017 A0A194QJ23 A0A2A4JJR5 A0A0L7LC11 A0A0L7LHM6 A0A194QJ80 A0A0L7KWB8 A0A2A4JT91 A0A0N1IG19 A0A212F9C9 A0A0N1IPS2 A0A0L7LCK3 H9J4X7 A0A0N1I9K7 A0A2A4IVM9 A0A194QJI7 A0A194QHU0 A0A023PNK9 A0A0N0PAH4 A0A194R9K8 A0A2A4JTG8 A0A2H1WVP2 A0A2W1BFI0 A0A2H1WB04 A0A2A4J748 B0W5I2 A0A2A4JNG3 A0A0N0PEP7 A0A0L7L7N1 A0A2H1WAU6 A0A194QEH9 A0A0L7KLX5 A0A212ENJ2 A0A0L7LIB6 A0A212F9B1 A0A194QPY2 A0A0N0PEE0 A0A212F161 A0A0N1IHN6 A0A2W1BMU2 A0A3S2LEF2 A0A336MDU0 A0A0N0PE20 A0A212FJ74 H9J525 H9J4X3 A0A194QJ30 A0A182GCQ0 A0A182GTB3 H9IV67 A0A2A4JUU3 A0A2W1BD78 H9J4X0 A0A0N1IDG3 A0A2H1W936 A0A194QJ40 Q17LS5
A0A2A4JBS9 H9JFD5 A0A2W1BR72 A0A2H1X3V1 A0A2H1X3Y1 A0A2W1BJK8 A0A2W1BLU7 A0A2W1BQ15 A0A2H1VQR9 A0A2H1VYW0 A0A2A4K454 A0A2A4K3Q9 A0A2H1VYX5 A0A0L7LBU2 A0A0L7KRG0 A0A0L7LW97 A0A0L7LSQ8 A0A194Q2H3 A0A212EJQ6 A0A194R9C4 A0A0N1IQT4 A0A212ETB8 A0A2H1WKZ9 A0A2A4KA42 A0A2W1B9E8 A0A2A4KB29 A0A2H1WK46 A0A2W1BN20 A0A2W1BF54 A0A2H1WX63 A0A2A4JKJ9 A0A194QKY0 H9J017 A0A194QJ23 A0A2A4JJR5 A0A0L7LC11 A0A0L7LHM6 A0A194QJ80 A0A0L7KWB8 A0A2A4JT91 A0A0N1IG19 A0A212F9C9 A0A0N1IPS2 A0A0L7LCK3 H9J4X7 A0A0N1I9K7 A0A2A4IVM9 A0A194QJI7 A0A194QHU0 A0A023PNK9 A0A0N0PAH4 A0A194R9K8 A0A2A4JTG8 A0A2H1WVP2 A0A2W1BFI0 A0A2H1WB04 A0A2A4J748 B0W5I2 A0A2A4JNG3 A0A0N0PEP7 A0A0L7L7N1 A0A2H1WAU6 A0A194QEH9 A0A0L7KLX5 A0A212ENJ2 A0A0L7LIB6 A0A212F9B1 A0A194QPY2 A0A0N0PEE0 A0A212F161 A0A0N1IHN6 A0A2W1BMU2 A0A3S2LEF2 A0A336MDU0 A0A0N0PE20 A0A212FJ74 H9J525 H9J4X3 A0A194QJ30 A0A182GCQ0 A0A182GTB3 H9IV67 A0A2A4JUU3 A0A2W1BD78 H9J4X0 A0A0N1IDG3 A0A2H1W936 A0A194QJ40 Q17LS5
PDB
4LDS
E-value=7.73476e-14,
Score=188
Ontologies
GO
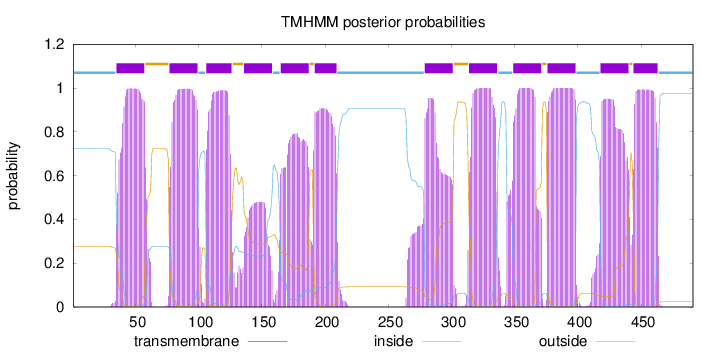
Topology
Length:
491
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
240.73144
Exp number, first 60 AAs:
21.95596
Total prob of N-in:
0.72273
POSSIBLE N-term signal
sequence
inside
1 - 34
TMhelix
35 - 57
outside
58 - 76
TMhelix
77 - 99
inside
100 - 105
TMhelix
106 - 126
outside
127 - 135
TMhelix
136 - 158
inside
159 - 164
TMhelix
165 - 187
outside
188 - 191
TMhelix
192 - 209
inside
210 - 278
TMhelix
279 - 301
outside
302 - 313
TMhelix
314 - 336
inside
337 - 348
TMhelix
349 - 371
outside
372 - 375
TMhelix
376 - 398
inside
399 - 417
TMhelix
418 - 440
outside
441 - 443
TMhelix
444 - 463
inside
464 - 491
Population Genetic Test Statistics
Pi
176.934265
Theta
176.468961
Tajima's D
0.386601
CLR
1.304153
CSRT
0.478526073696315
Interpretation
Uncertain
