Gene
KWMTBOMO06186
Annotation
PREDICTED:_zinc_finger_protein_28-like_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 3.248
Sequence
CDS
ATGAGTGCACGAGATCACGGCCCTCCTGGTGTTGAGTGGTCACCGGAGCCCATGGACATCAACAACCTCCTCATATCGAAGTGGCGTCTCAAGGACCACGAGAGGAAGCACCACGGGTCGAGCCGACCCAGGGACCACTGCTGCAAGACCTGCGGGAAGAGATTCTACACTCTATCCAGCCTGCACGGTCATCAGTTGATCCACGACCCGCAGAGGGGCTACATGTGCGAGGAGTGCGGGGACACGTTCAAACAGCCGGCCGCTCTCTATACGCATTCTAGATTGGTGCACAGGAATGCGAAGTGA
Protein
MSARDHGPPGVEWSPEPMDINNLLISKWRLKDHERKHHGSSRPRDHCCKTCGKRFYTLSSLHGHQLIHDPQRGYMCEECGDTFKQPAALYTHSRLVHRNAK
Summary
Uniprot
A0A3S2NV19
A0A0N0PBL1
A0A0N0PAK7
A0A2A4J7P7
A0A2H1WUB2
A0A2W1B042
+ More
A0A1U7QYU9 A0A060YK24 M0RCK0 Q5PPQ4 Q3U2V8 A0A3P8YVU8 A0A1E1W6H5 Q80UY3 A0A212F8F4 A0A1B0CYR2 A0A3Q2UZH3 A0A3B5LTZ6 G3IM67 A0A1S3QKK8 H9IWZ5 S4NN42 A0A3S2PCA5 H9H749 A0A3Q2V817 A0A2H1VWX0 A0A1B6GKV0 H9IWM4 W5LWU9 G3VJX4 A0A182T8W5 A0A3B3H7H6 A0A060WFS9 A0A1S3WKQ2 S4PXE3 A0A2W1BCQ2 A0A3P9KID7 A0A3B3INK9 W5LWY6 A0A1S3RLF0 H9JQV0 A0A1S3QMN9 A0A3Q0J1R5 A0A2G9QI59 A0A3P9M1F6 A0A2R7VY71 S4RFV1 G1KVR1 A0A3P9KIN5 S4RFV3 H0ZW53 Q4TAD0 A0A0N1PHY6 A0A0N1IEI9 A0A087UAB4
A0A1U7QYU9 A0A060YK24 M0RCK0 Q5PPQ4 Q3U2V8 A0A3P8YVU8 A0A1E1W6H5 Q80UY3 A0A212F8F4 A0A1B0CYR2 A0A3Q2UZH3 A0A3B5LTZ6 G3IM67 A0A1S3QKK8 H9IWZ5 S4NN42 A0A3S2PCA5 H9H749 A0A3Q2V817 A0A2H1VWX0 A0A1B6GKV0 H9IWM4 W5LWU9 G3VJX4 A0A182T8W5 A0A3B3H7H6 A0A060WFS9 A0A1S3WKQ2 S4PXE3 A0A2W1BCQ2 A0A3P9KID7 A0A3B3INK9 W5LWY6 A0A1S3RLF0 H9JQV0 A0A1S3QMN9 A0A3Q0J1R5 A0A2G9QI59 A0A3P9M1F6 A0A2R7VY71 S4RFV1 G1KVR1 A0A3P9KIN5 S4RFV3 H0ZW53 Q4TAD0 A0A0N1PHY6 A0A0N1IEI9 A0A087UAB4
Pubmed
EMBL
RSAL01000063
RVE49479.1
KQ460969
KPJ10206.1
KQ458725
KPJ05348.1
+ More
NWSH01002684 PCG67688.1 ODYU01010720 SOQ56004.1 KZ150535 PZC70589.1 FR912482 CDQ91922.1 AABR07003234 AC154108 BC087557 AAH87557.1 AK155078 BAE33032.1 GDQN01008479 JAT82575.1 BC043301 AAH43301.1 AGBW02009729 OWR50026.1 AJVK01020326 AJVK01020327 JH004522 EGW05050.1 BABH01021157 GAIX01014096 JAA78464.1 RSAL01000105 RVE47324.1 ODYU01004954 SOQ45347.1 GECZ01006836 JAS62933.1 BABH01021167 AEFK01123826 FR904531 CDQ66133.1 GAIX01005188 JAA87372.1 KZ150132 PZC73042.1 BABH01028690 KV990864 PIO14763.1 KK854082 PTY10885.1 ABQF01073408 ABQF01073409 CAAE01007378 CAF90152.1 KQ460555 KPJ14020.1 KQ458486 KPJ05869.1 KK118972 KFM74303.1
NWSH01002684 PCG67688.1 ODYU01010720 SOQ56004.1 KZ150535 PZC70589.1 FR912482 CDQ91922.1 AABR07003234 AC154108 BC087557 AAH87557.1 AK155078 BAE33032.1 GDQN01008479 JAT82575.1 BC043301 AAH43301.1 AGBW02009729 OWR50026.1 AJVK01020326 AJVK01020327 JH004522 EGW05050.1 BABH01021157 GAIX01014096 JAA78464.1 RSAL01000105 RVE47324.1 ODYU01004954 SOQ45347.1 GECZ01006836 JAS62933.1 BABH01021167 AEFK01123826 FR904531 CDQ66133.1 GAIX01005188 JAA87372.1 KZ150132 PZC73042.1 BABH01028690 KV990864 PIO14763.1 KK854082 PTY10885.1 ABQF01073408 ABQF01073409 CAAE01007378 CAF90152.1 KQ460555 KPJ14020.1 KQ458486 KPJ05869.1 KK118972 KFM74303.1
Proteomes
UP000283053
UP000053240
UP000053268
UP000218220
UP000189706
UP000193380
+ More
UP000002494 UP000000589 UP000265140 UP000007151 UP000092462 UP000264840 UP000261380 UP000001075 UP000087266 UP000005204 UP000002280 UP000018468 UP000007648 UP000075901 UP000001038 UP000079721 UP000265180 UP000079169 UP000245300 UP000001646 UP000007754 UP000054359
UP000002494 UP000000589 UP000265140 UP000007151 UP000092462 UP000264840 UP000261380 UP000001075 UP000087266 UP000005204 UP000002280 UP000018468 UP000007648 UP000075901 UP000001038 UP000079721 UP000265180 UP000079169 UP000245300 UP000001646 UP000007754 UP000054359
Interpro
ProteinModelPortal
A0A3S2NV19
A0A0N0PBL1
A0A0N0PAK7
A0A2A4J7P7
A0A2H1WUB2
A0A2W1B042
+ More
A0A1U7QYU9 A0A060YK24 M0RCK0 Q5PPQ4 Q3U2V8 A0A3P8YVU8 A0A1E1W6H5 Q80UY3 A0A212F8F4 A0A1B0CYR2 A0A3Q2UZH3 A0A3B5LTZ6 G3IM67 A0A1S3QKK8 H9IWZ5 S4NN42 A0A3S2PCA5 H9H749 A0A3Q2V817 A0A2H1VWX0 A0A1B6GKV0 H9IWM4 W5LWU9 G3VJX4 A0A182T8W5 A0A3B3H7H6 A0A060WFS9 A0A1S3WKQ2 S4PXE3 A0A2W1BCQ2 A0A3P9KID7 A0A3B3INK9 W5LWY6 A0A1S3RLF0 H9JQV0 A0A1S3QMN9 A0A3Q0J1R5 A0A2G9QI59 A0A3P9M1F6 A0A2R7VY71 S4RFV1 G1KVR1 A0A3P9KIN5 S4RFV3 H0ZW53 Q4TAD0 A0A0N1PHY6 A0A0N1IEI9 A0A087UAB4
A0A1U7QYU9 A0A060YK24 M0RCK0 Q5PPQ4 Q3U2V8 A0A3P8YVU8 A0A1E1W6H5 Q80UY3 A0A212F8F4 A0A1B0CYR2 A0A3Q2UZH3 A0A3B5LTZ6 G3IM67 A0A1S3QKK8 H9IWZ5 S4NN42 A0A3S2PCA5 H9H749 A0A3Q2V817 A0A2H1VWX0 A0A1B6GKV0 H9IWM4 W5LWU9 G3VJX4 A0A182T8W5 A0A3B3H7H6 A0A060WFS9 A0A1S3WKQ2 S4PXE3 A0A2W1BCQ2 A0A3P9KID7 A0A3B3INK9 W5LWY6 A0A1S3RLF0 H9JQV0 A0A1S3QMN9 A0A3Q0J1R5 A0A2G9QI59 A0A3P9M1F6 A0A2R7VY71 S4RFV1 G1KVR1 A0A3P9KIN5 S4RFV3 H0ZW53 Q4TAD0 A0A0N1PHY6 A0A0N1IEI9 A0A087UAB4
PDB
2I13
E-value=3.0867e-05,
Score=106
Ontologies
GO
Topology
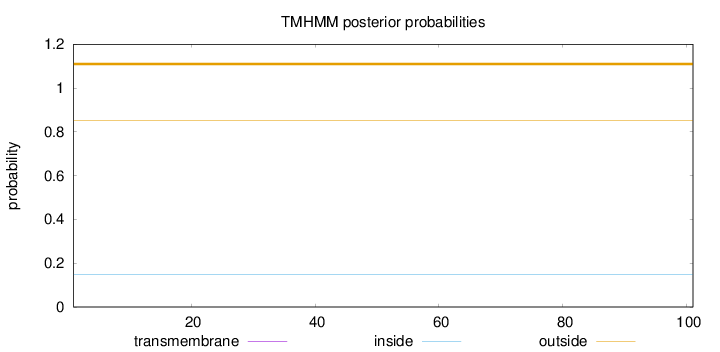
Length:
101
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.14896
outside
1 - 101
Population Genetic Test Statistics
Pi
276.409531
Theta
169.230898
Tajima's D
2.442941
CLR
0.668419
CSRT
0.938453077346133
Interpretation
Uncertain
