Pre Gene Modal
BGIBMGA001664
Annotation
glucose_transporter_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.883
Sequence
CDS
ATGAACCCGCGTCTCGCTTTCGCCATTATATCGTCGACCTGCTGGTCTGCCTTCCAGCATGGATACAATACAGGCGTGATAAACGCTCCGCAGGCGGTCATGAGCGAATGGCTTCAGAAAGAGGCGCTTAGTGGGACGAACACCTCGGTCTCCGCCATGGTCGATCCAAAGGTGACTTCGGTCTGGTCGGTGGCTGTCTCCATATACTGCGCCGGCGGCATGATTGGTGGTGTGATCACCGGAATCATTGCCGACAGATTCGGCAGAAAAGGCGGTCTGCTGCTCAACAACGTGCTGGTATTCCTGGCGGCCGCTCTGCAGGGAGCTTCGAAGGTCGTCAAATCGGCGGAGATGCTCATCCTTGGAAGATTACTCATTGGCGTTAACAGTGGCCTGAACGCGGGCTTAGTGCCTCTCTACCTGTCAGAGATATCGCCAGTGTCAATCAGAGGCTCGATCGGCACAGTTTACCAGCTCGTCATAACAGTCACGATACTGCTGTCCCAGCTGCTAGGGCTGAGCAGTGTGCTCGGCACGGAGGACGGCTGGCCATGGTTATTGGCACTGACAGCGGCGCCGGCCGTCCTGCAGTGTCTCACGCTGCCCCTATGCCCAGAATCGCCAAAGTACTTACTCCTGAACCAGGGTCGAGAGCTGCACGCCCAACGAGCATTGAACTGGCTGCGAGGCGACGTCGCGGTGCACGGCGAGATGGAAGAAATGCATCAGGAAGCGGAGAAGAATAAGATCAGCAAGAAGGTCACCCTGCGAGAGTTGTTCCGAAACAGGAACCTCCGTCTGCCCCTCTTCATATCCACAGTCGTGATGATAGCTCAACAGCTCTCCGGGATAAACGCTATAATATACTTTTCGACTGACATCTTTGAGAAGACTCATCTCGGATCTGATGCTGCGCAGTACGCTACTTTGGGCATAGGCGCCATGAATGTCGTAATGACGATAGCCAGCCTCGTGCTCGTCGAGGTGGCTGGCAGAAAGACCCTCCTGCTGGCAGGGTTCTCGGGAATGTTCATATGTACCGTCGGCATAACAGTCGCTACTTCATACACTCAGCACGTCTGGGTGTCGTATCTGTGCATAGCGCTCGTGGTTCTCTTCGTGACGATGTTCGCGATCGGTCCGGGTTCGATACCCTGGTTCTTGGTGACAGAGTTATTCAACCAATCATCTCGGCCGGCCGCGTCATCTGTCGCTGTCACTGTCAATTGGACGGCCAATTTCATTGTCGGATTAAGCTTCTTACCGCTCTCGCTCGCCCTGGGGAACAATGTTTTCGTGATATTCGCGGTCCTACAATTCCTGTTCATAATTTTTATTTACACTAAAGTACCTGAGACGAAAAATAAGACTGTTGACGAGATAACCGCCATGTTTAGGCAGCGGATGTAG
Protein
MNPRLAFAIISSTCWSAFQHGYNTGVINAPQAVMSEWLQKEALSGTNTSVSAMVDPKVTSVWSVAVSIYCAGGMIGGVITGIIADRFGRKGGLLLNNVLVFLAAALQGASKVVKSAEMLILGRLLIGVNSGLNAGLVPLYLSEISPVSIRGSIGTVYQLVITVTILLSQLLGLSSVLGTEDGWPWLLALTAAPAVLQCLTLPLCPESPKYLLLNQGRELHAQRALNWLRGDVAVHGEMEEMHQEAEKNKISKKVTLRELFRNRNLRLPLFISTVVMIAQQLSGINAIIYFSTDIFEKTHLGSDAAQYATLGIGAMNVVMTIASLVLVEVAGRKTLLLAGFSGMFICTVGITVATSYTQHVWVSYLCIALVVLFVTMFAIGPGSIPWFLVTELFNQSSRPAASSVAVTVNWTANFIVGLSFLPLSLALGNNVFVIFAVLQFLFIIFIYTKVPETKNKTVDEITAMFRQRM
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9IWN2
E3UVB5
A0A1W6S010
A0A2H1W5S7
A0A0N0PA92
A0A212EUJ6
+ More
A0A3S2LAP1 A0A1S3CV23 A0A1S3CWD1 A0A2P8XEX6 A0A026WW63 A0A2J7RR30 A0A2J7RR22 A0A2J7RR32 E0VJK9 A0A2J7RR20 A0A195FTS9 A0A2J7RR25 A0A195BC83 A0A2J7RR24 A0A1Z1XG24 A0A151XAA9 A0A195C803 D6WL90 A0A158P3Y9 A0A0J7KFE0 A0A154PA49 V9IAG4 V9IBR3 A0A0L7RDM7 A0A088ART6 D4AHX5 A0A2A3E2E6 V9ICA0 A0A1B6LPQ6 A0A067RNH1 A0A1B6C3D1 E9J2L5 A0A2S2Q6S7 A0A1B6EH05 A0A1W4XK78 A0A0C9R8Y6 A0A0C9QZU0 A0A0C9Q5L6 A0A1B6D994 A0A1W4X9T1 A0A0M8ZM67 A0A0C9RQB2 A0A1B6K0X7 A0A2H8TQK4 W0FVI8 A0A1B6GVS8 W0FQS9 E2A2Q5 E2APZ0 A0A195DCI6 A0A3L8DI80 A0A310SI35 A0A2P2I4R0 J9JP06 A0A0H3XSL9 A0A023F563 A0A226EN11 A0A2P6KTM4 E9FYK1 F4WM42 B7QM62 A0A0P5ZMG6 A0A0P6JT67 A0A131XGB3 A0A0T6ATB0 A0A147BTG0 A0A0P6CI00 A0A1Y1LLG5 A0A2R5LM20 A0A023GKZ1 A0A224XC28 K7J6Z2 L7M3M0 A0A224Z582 A0A146LZR5 A0A131Z0A0 A0A146L6F7 A0A1Z5KVC6 A0A1B6F454 A0A2S2NM67 A0A087TJW5 N6TBY5 A0A2P1JJ71 T1HKP2 E2BX45 A0A293N3H5 A0A1B6F6N4 A0A087ULC1 D4AHW8 A0A1D2MR33 V5ICR0 A0A0P5FD17
A0A3S2LAP1 A0A1S3CV23 A0A1S3CWD1 A0A2P8XEX6 A0A026WW63 A0A2J7RR30 A0A2J7RR22 A0A2J7RR32 E0VJK9 A0A2J7RR20 A0A195FTS9 A0A2J7RR25 A0A195BC83 A0A2J7RR24 A0A1Z1XG24 A0A151XAA9 A0A195C803 D6WL90 A0A158P3Y9 A0A0J7KFE0 A0A154PA49 V9IAG4 V9IBR3 A0A0L7RDM7 A0A088ART6 D4AHX5 A0A2A3E2E6 V9ICA0 A0A1B6LPQ6 A0A067RNH1 A0A1B6C3D1 E9J2L5 A0A2S2Q6S7 A0A1B6EH05 A0A1W4XK78 A0A0C9R8Y6 A0A0C9QZU0 A0A0C9Q5L6 A0A1B6D994 A0A1W4X9T1 A0A0M8ZM67 A0A0C9RQB2 A0A1B6K0X7 A0A2H8TQK4 W0FVI8 A0A1B6GVS8 W0FQS9 E2A2Q5 E2APZ0 A0A195DCI6 A0A3L8DI80 A0A310SI35 A0A2P2I4R0 J9JP06 A0A0H3XSL9 A0A023F563 A0A226EN11 A0A2P6KTM4 E9FYK1 F4WM42 B7QM62 A0A0P5ZMG6 A0A0P6JT67 A0A131XGB3 A0A0T6ATB0 A0A147BTG0 A0A0P6CI00 A0A1Y1LLG5 A0A2R5LM20 A0A023GKZ1 A0A224XC28 K7J6Z2 L7M3M0 A0A224Z582 A0A146LZR5 A0A131Z0A0 A0A146L6F7 A0A1Z5KVC6 A0A1B6F454 A0A2S2NM67 A0A087TJW5 N6TBY5 A0A2P1JJ71 T1HKP2 E2BX45 A0A293N3H5 A0A1B6F6N4 A0A087ULC1 D4AHW8 A0A1D2MR33 V5ICR0 A0A0P5FD17
Pubmed
EMBL
BABH01021144
HM593515
ADP37708.1
KY197474
ARO71211.1
ODYU01006492
+ More
SOQ48373.1 KQ458725 KPJ05347.1 AGBW02012366 OWR45159.1 RSAL01000063 RVE49480.1 PYGN01002411 PSN30545.1 KK107109 EZA59364.1 NEVH01000611 PNF43283.1 PNF43281.1 PNF43285.1 DS235225 EEB13565.1 PNF43282.1 KQ981264 KYN44025.1 PNF43286.1 KQ976519 KYM82158.1 PNF43284.1 KY350172 ARX98211.1 KQ982353 KYQ57208.1 KQ978220 KYM96301.1 KQ971343 EFA03487.1 ADTU01001343 ADTU01001344 LBMM01008302 KMQ88997.1 KQ434856 KZC08711.1 JR037992 AEY58065.1 JR037994 AEY58067.1 KQ414614 KOC68836.1 AB550003 BAI83424.1 KZ288427 PBC25870.1 JR037993 AEY58066.1 GEBQ01022372 GEBQ01014309 JAT17605.1 JAT25668.1 KK852521 KDR22120.1 GEDC01029276 JAS08022.1 GL767895 EFZ12945.1 GGMS01004098 MBY73301.1 GEDC01000093 JAS37205.1 GBYB01009287 JAG79054.1 GBYB01009289 JAG79056.1 GBYB01009383 JAG79150.1 GEDC01015133 JAS22165.1 KQ438551 KOX67160.1 GBYB01009386 JAG79153.1 GECU01002587 JAT05120.1 GFXV01004620 MBW16425.1 KF803263 AHF27417.1 GECZ01003250 JAS66519.1 KF803264 AHF27418.1 GL436203 EFN72280.1 GL441645 EFN64477.1 KQ980989 KYN10598.1 QOIP01000008 RLU19629.1 KQ760869 OAD58743.1 IACF01003401 LAB69017.1 ABLF02029594 ABLF02029596 ABLF02029598 ABLF02047180 KR047102 AKM95032.1 GBBI01002016 JAC16696.1 LNIX01000003 OXA58588.1 MWRG01005420 PRD29662.1 GL732527 EFX87752.1 GL888217 EGI64686.1 ABJB010969653 ABJB011094387 DS969311 EEC19934.1 GDIQ01210261 GDIP01041843 LRGB01001151 JAK41464.1 JAM61872.1 KZS13476.1 GDIQ01009392 JAN85345.1 GEFH01002532 JAP66049.1 LJIG01022875 KRT78281.1 GEGO01001377 JAR94027.1 GDIP01008328 JAM95387.1 GEZM01055074 JAV73170.1 GGLE01006436 MBY10562.1 GBBM01000924 JAC34494.1 GFTR01006450 JAW09976.1 GACK01007296 JAA57738.1 GFPF01012113 MAA23259.1 GDHC01015702 GDHC01008865 GDHC01006779 JAQ02927.1 JAQ09764.1 JAQ11850.1 GEDV01004661 JAP83896.1 GDHC01015877 GDHC01003860 JAQ02752.1 JAQ14769.1 GFJQ02007995 JAV98974.1 GECZ01024814 JAS44955.1 GGMR01005257 MBY17876.1 KK115557 KFM65404.1 APGK01043875 KB741018 KB632152 ENN75243.1 ERL89130.1 MF385066 AVN99061.1 ACPB03000923 GL451202 EFN79764.1 GFWV01022212 MAA46939.1 GECZ01023909 JAS45860.1 KK120376 KFM78160.1 AB549996 BAI83417.1 LJIJ01000693 ODM95264.1 GANP01013252 JAB71216.1 GDIQ01259982 JAJ91742.1
SOQ48373.1 KQ458725 KPJ05347.1 AGBW02012366 OWR45159.1 RSAL01000063 RVE49480.1 PYGN01002411 PSN30545.1 KK107109 EZA59364.1 NEVH01000611 PNF43283.1 PNF43281.1 PNF43285.1 DS235225 EEB13565.1 PNF43282.1 KQ981264 KYN44025.1 PNF43286.1 KQ976519 KYM82158.1 PNF43284.1 KY350172 ARX98211.1 KQ982353 KYQ57208.1 KQ978220 KYM96301.1 KQ971343 EFA03487.1 ADTU01001343 ADTU01001344 LBMM01008302 KMQ88997.1 KQ434856 KZC08711.1 JR037992 AEY58065.1 JR037994 AEY58067.1 KQ414614 KOC68836.1 AB550003 BAI83424.1 KZ288427 PBC25870.1 JR037993 AEY58066.1 GEBQ01022372 GEBQ01014309 JAT17605.1 JAT25668.1 KK852521 KDR22120.1 GEDC01029276 JAS08022.1 GL767895 EFZ12945.1 GGMS01004098 MBY73301.1 GEDC01000093 JAS37205.1 GBYB01009287 JAG79054.1 GBYB01009289 JAG79056.1 GBYB01009383 JAG79150.1 GEDC01015133 JAS22165.1 KQ438551 KOX67160.1 GBYB01009386 JAG79153.1 GECU01002587 JAT05120.1 GFXV01004620 MBW16425.1 KF803263 AHF27417.1 GECZ01003250 JAS66519.1 KF803264 AHF27418.1 GL436203 EFN72280.1 GL441645 EFN64477.1 KQ980989 KYN10598.1 QOIP01000008 RLU19629.1 KQ760869 OAD58743.1 IACF01003401 LAB69017.1 ABLF02029594 ABLF02029596 ABLF02029598 ABLF02047180 KR047102 AKM95032.1 GBBI01002016 JAC16696.1 LNIX01000003 OXA58588.1 MWRG01005420 PRD29662.1 GL732527 EFX87752.1 GL888217 EGI64686.1 ABJB010969653 ABJB011094387 DS969311 EEC19934.1 GDIQ01210261 GDIP01041843 LRGB01001151 JAK41464.1 JAM61872.1 KZS13476.1 GDIQ01009392 JAN85345.1 GEFH01002532 JAP66049.1 LJIG01022875 KRT78281.1 GEGO01001377 JAR94027.1 GDIP01008328 JAM95387.1 GEZM01055074 JAV73170.1 GGLE01006436 MBY10562.1 GBBM01000924 JAC34494.1 GFTR01006450 JAW09976.1 GACK01007296 JAA57738.1 GFPF01012113 MAA23259.1 GDHC01015702 GDHC01008865 GDHC01006779 JAQ02927.1 JAQ09764.1 JAQ11850.1 GEDV01004661 JAP83896.1 GDHC01015877 GDHC01003860 JAQ02752.1 JAQ14769.1 GFJQ02007995 JAV98974.1 GECZ01024814 JAS44955.1 GGMR01005257 MBY17876.1 KK115557 KFM65404.1 APGK01043875 KB741018 KB632152 ENN75243.1 ERL89130.1 MF385066 AVN99061.1 ACPB03000923 GL451202 EFN79764.1 GFWV01022212 MAA46939.1 GECZ01023909 JAS45860.1 KK120376 KFM78160.1 AB549996 BAI83417.1 LJIJ01000693 ODM95264.1 GANP01013252 JAB71216.1 GDIQ01259982 JAJ91742.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000283053
UP000079169
UP000245037
+ More
UP000053097 UP000235965 UP000009046 UP000078541 UP000078540 UP000075809 UP000078542 UP000007266 UP000005205 UP000036403 UP000076502 UP000053825 UP000005203 UP000242457 UP000027135 UP000192223 UP000053105 UP000000311 UP000078492 UP000279307 UP000007819 UP000198287 UP000000305 UP000007755 UP000001555 UP000076858 UP000002358 UP000054359 UP000019118 UP000030742 UP000015103 UP000008237 UP000094527
UP000053097 UP000235965 UP000009046 UP000078541 UP000078540 UP000075809 UP000078542 UP000007266 UP000005205 UP000036403 UP000076502 UP000053825 UP000005203 UP000242457 UP000027135 UP000192223 UP000053105 UP000000311 UP000078492 UP000279307 UP000007819 UP000198287 UP000000305 UP000007755 UP000001555 UP000076858 UP000002358 UP000054359 UP000019118 UP000030742 UP000015103 UP000008237 UP000094527
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9IWN2
E3UVB5
A0A1W6S010
A0A2H1W5S7
A0A0N0PA92
A0A212EUJ6
+ More
A0A3S2LAP1 A0A1S3CV23 A0A1S3CWD1 A0A2P8XEX6 A0A026WW63 A0A2J7RR30 A0A2J7RR22 A0A2J7RR32 E0VJK9 A0A2J7RR20 A0A195FTS9 A0A2J7RR25 A0A195BC83 A0A2J7RR24 A0A1Z1XG24 A0A151XAA9 A0A195C803 D6WL90 A0A158P3Y9 A0A0J7KFE0 A0A154PA49 V9IAG4 V9IBR3 A0A0L7RDM7 A0A088ART6 D4AHX5 A0A2A3E2E6 V9ICA0 A0A1B6LPQ6 A0A067RNH1 A0A1B6C3D1 E9J2L5 A0A2S2Q6S7 A0A1B6EH05 A0A1W4XK78 A0A0C9R8Y6 A0A0C9QZU0 A0A0C9Q5L6 A0A1B6D994 A0A1W4X9T1 A0A0M8ZM67 A0A0C9RQB2 A0A1B6K0X7 A0A2H8TQK4 W0FVI8 A0A1B6GVS8 W0FQS9 E2A2Q5 E2APZ0 A0A195DCI6 A0A3L8DI80 A0A310SI35 A0A2P2I4R0 J9JP06 A0A0H3XSL9 A0A023F563 A0A226EN11 A0A2P6KTM4 E9FYK1 F4WM42 B7QM62 A0A0P5ZMG6 A0A0P6JT67 A0A131XGB3 A0A0T6ATB0 A0A147BTG0 A0A0P6CI00 A0A1Y1LLG5 A0A2R5LM20 A0A023GKZ1 A0A224XC28 K7J6Z2 L7M3M0 A0A224Z582 A0A146LZR5 A0A131Z0A0 A0A146L6F7 A0A1Z5KVC6 A0A1B6F454 A0A2S2NM67 A0A087TJW5 N6TBY5 A0A2P1JJ71 T1HKP2 E2BX45 A0A293N3H5 A0A1B6F6N4 A0A087ULC1 D4AHW8 A0A1D2MR33 V5ICR0 A0A0P5FD17
A0A3S2LAP1 A0A1S3CV23 A0A1S3CWD1 A0A2P8XEX6 A0A026WW63 A0A2J7RR30 A0A2J7RR22 A0A2J7RR32 E0VJK9 A0A2J7RR20 A0A195FTS9 A0A2J7RR25 A0A195BC83 A0A2J7RR24 A0A1Z1XG24 A0A151XAA9 A0A195C803 D6WL90 A0A158P3Y9 A0A0J7KFE0 A0A154PA49 V9IAG4 V9IBR3 A0A0L7RDM7 A0A088ART6 D4AHX5 A0A2A3E2E6 V9ICA0 A0A1B6LPQ6 A0A067RNH1 A0A1B6C3D1 E9J2L5 A0A2S2Q6S7 A0A1B6EH05 A0A1W4XK78 A0A0C9R8Y6 A0A0C9QZU0 A0A0C9Q5L6 A0A1B6D994 A0A1W4X9T1 A0A0M8ZM67 A0A0C9RQB2 A0A1B6K0X7 A0A2H8TQK4 W0FVI8 A0A1B6GVS8 W0FQS9 E2A2Q5 E2APZ0 A0A195DCI6 A0A3L8DI80 A0A310SI35 A0A2P2I4R0 J9JP06 A0A0H3XSL9 A0A023F563 A0A226EN11 A0A2P6KTM4 E9FYK1 F4WM42 B7QM62 A0A0P5ZMG6 A0A0P6JT67 A0A131XGB3 A0A0T6ATB0 A0A147BTG0 A0A0P6CI00 A0A1Y1LLG5 A0A2R5LM20 A0A023GKZ1 A0A224XC28 K7J6Z2 L7M3M0 A0A224Z582 A0A146LZR5 A0A131Z0A0 A0A146L6F7 A0A1Z5KVC6 A0A1B6F454 A0A2S2NM67 A0A087TJW5 N6TBY5 A0A2P1JJ71 T1HKP2 E2BX45 A0A293N3H5 A0A1B6F6N4 A0A087ULC1 D4AHW8 A0A1D2MR33 V5ICR0 A0A0P5FD17
PDB
5EQI
E-value=2.59609e-91,
Score=856
Ontologies
GO
Topology
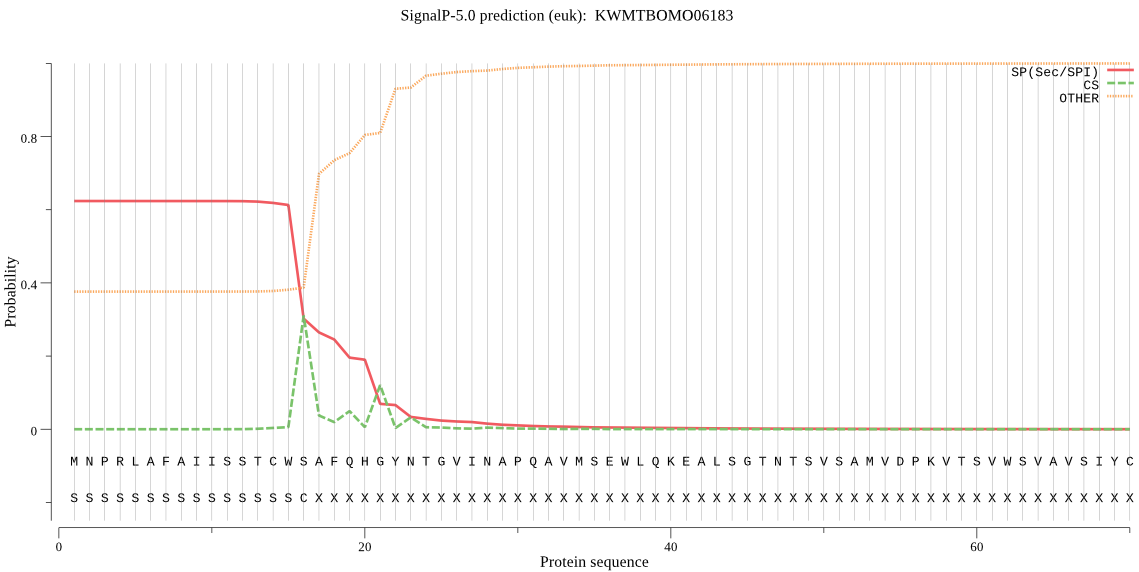
SignalP
Position: 1 - 16,
Likelihood: 0.623239
Length:
469
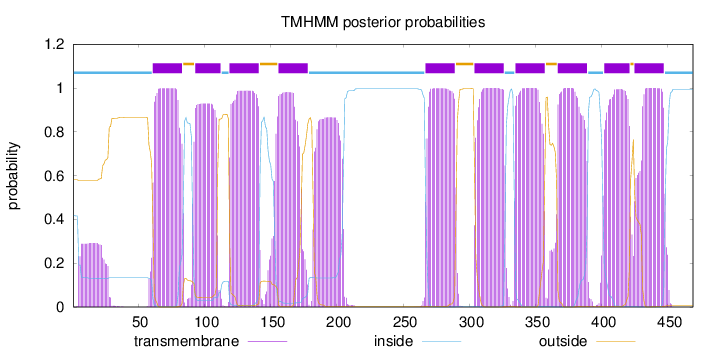
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
243.08075
Exp number, first 60 AAs:
6.75531
Total prob of N-in:
0.41861
inside
1 - 60
TMhelix
61 - 83
outside
84 - 92
TMhelix
93 - 112
inside
113 - 118
TMhelix
119 - 141
outside
142 - 155
TMhelix
156 - 178
inside
179 - 266
TMhelix
267 - 289
outside
290 - 303
TMhelix
304 - 326
inside
327 - 334
TMhelix
335 - 357
outside
358 - 366
TMhelix
367 - 389
inside
390 - 401
TMhelix
402 - 421
outside
422 - 424
TMhelix
425 - 447
inside
448 - 469
Population Genetic Test Statistics
Pi
266.012383
Theta
158.864836
Tajima's D
2.763339
CLR
0.238535
CSRT
0.967551622418879
Interpretation
Uncertain
