Gene
KWMTBOMO06179
Pre Gene Modal
BGIBMGA001666
Annotation
PREDICTED:_nucleosome-remodeling_factor_subunit_BPTF_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 4.807
Sequence
CDS
ATGCTTTCCGTTTCGCAGTTGAGCAGAGAAGAGGAGAAGGACGGAAAGAAAAAGAGAGACTATTCTAGAAAGAAGAAATCCGATTCTCGAACCTGCTCAGGGTCGGAGAGCGCTGCGGAGACCAGCGCCCCGGACTCGCCCAGCGGCAACGACGCCGAGAGACAGCACCGCCTGTGGAGGAAGTCCGTCATGCTGGTCTACAGCAGATTGTGCGCCCATAAATACGCTTCGTTGTTCCTCCGTCCGATCAGCGACGAGGAGGCGCCCGGGTACAGCGTCGTCGTGAAGCGGCCCATGGACCTCACCACGATCCGCAGGAACATCGACAACGGAAACATCAGGACGACGGCCGAGTTCCAGCGCGACGTGCTCCTGATGCTGTCCAATGCCCTCATCTACAACAGCACGGAGCACAGTGTCCACTCGATGGCGAAGGAGATGCACGAGGAGGCTCAGTGTCAGCTGGGCATGCTGGTGGCTGCGCAGGCGCACGCCGGACTGTGCGCCGCCCCCCCGCTGCGGAGGAAGAGGAGGCGCCACCACTCCCCCCTGCACCACCCCGCGCACTACACGCGCCCGCACTGA
Protein
MLSVSQLSREEEKDGKKKRDYSRKKKSDSRTCSGSESAAETSAPDSPSGNDAERQHRLWRKSVMLVYSRLCAHKYASLFLRPISDEEAPGYSVVVKRPMDLTTIRRNIDNGNIRTTAEFQRDVLLMLSNALIYNSTEHSVHSMAKEMHEEAQCQLGMLVAAQAHAGLCAAPPLRRKRRRHHSPLHHPAHYTRPH
Summary
Uniprot
H9IWN4
A0A2A4K5G6
A0A212F487
S4PGH2
A0A1E1WLI2
A0A194Q4Z9
+ More
V5I6E6 E0VFW5 A0A067R8G4 A0A3S3PA68 A0A0P5HU95 A0A0P5HXK0 A0A0B7AN89 T1KG49 A0A026W9H9 A0A3L8DYM5 A0A3S1AXJ8 A0A210QL55 A0A3R7M2X0 A0A0B7AQV6 A0A0B7AQU7 A0A195BRB2 A0A1E1XJS8 A0A158NZL7 T1IIC5 T1FUX7 A0A0P4XX32 A0A0P5AH37 A0A0B7AQT4 A0A0P6J8B5 A0A0P5YIK9 A0A0P5PH60 A0A131Z0S7 A0A1Z5L3V3 E9G2D3 A0A0B7AMM0 A0A0L8GJP8 A0A224YP87 A0A224YSJ0 A0A0B7ANX6 A0A0B7AMP1 A0A1W4XC19 N6TAA1 E7F3M6 W5LHN2
V5I6E6 E0VFW5 A0A067R8G4 A0A3S3PA68 A0A0P5HU95 A0A0P5HXK0 A0A0B7AN89 T1KG49 A0A026W9H9 A0A3L8DYM5 A0A3S1AXJ8 A0A210QL55 A0A3R7M2X0 A0A0B7AQV6 A0A0B7AQU7 A0A195BRB2 A0A1E1XJS8 A0A158NZL7 T1IIC5 T1FUX7 A0A0P4XX32 A0A0P5AH37 A0A0B7AQT4 A0A0P6J8B5 A0A0P5YIK9 A0A0P5PH60 A0A131Z0S7 A0A1Z5L3V3 E9G2D3 A0A0B7AMM0 A0A0L8GJP8 A0A224YP87 A0A224YSJ0 A0A0B7ANX6 A0A0B7AMP1 A0A1W4XC19 N6TAA1 E7F3M6 W5LHN2
Pubmed
EMBL
BABH01021142
BABH01021143
NWSH01000107
PCG79471.1
AGBW02010415
OWR48547.1
+ More
GAIX01000764 JAA91796.1 GDQN01003190 JAT87864.1 KQ459463 KPJ00434.1 GALX01008320 JAB60146.1 DS235124 EEB12271.1 KK852669 KDR18841.1 NCKU01001805 RWS11201.1 GDIQ01222199 JAK29526.1 GDIQ01222200 JAK29525.1 HACG01035448 CEK82313.1 CAEY01000044 KK107321 EZA52700.1 QOIP01000002 RLU25392.1 RQTK01001561 RUS69833.1 NEDP02003088 OWF49477.1 QCYY01002437 ROT70365.1 HACG01035450 CEK82315.1 HACG01035440 CEK82305.1 KQ976423 KYM88749.1 GFAA01004163 JAT99271.1 ADTU01000888 AFFK01014253 AMQM01006515 AMQM01006516 KB097487 ESN96827.1 GDIP01235564 GDIP01038984 LRGB01000512 JAI87837.1 KZS18695.1 GDIP01199668 JAJ23734.1 HACG01035425 CEK82290.1 GDIQ01011400 JAN83337.1 GDIP01058789 JAM44926.1 GDIQ01138724 JAL13002.1 GEDV01004491 JAP84066.1 GFJQ02005092 JAW01878.1 GL732530 EFX86231.1 HACG01035419 CEK82284.1 KQ421523 KOF77227.1 GFPF01006413 MAA17559.1 GFPF01006415 MAA17561.1 HACG01035442 CEK82307.1 HACG01035439 CEK82304.1 APGK01045628 KB741037 ENN74658.1 BX510945
GAIX01000764 JAA91796.1 GDQN01003190 JAT87864.1 KQ459463 KPJ00434.1 GALX01008320 JAB60146.1 DS235124 EEB12271.1 KK852669 KDR18841.1 NCKU01001805 RWS11201.1 GDIQ01222199 JAK29526.1 GDIQ01222200 JAK29525.1 HACG01035448 CEK82313.1 CAEY01000044 KK107321 EZA52700.1 QOIP01000002 RLU25392.1 RQTK01001561 RUS69833.1 NEDP02003088 OWF49477.1 QCYY01002437 ROT70365.1 HACG01035450 CEK82315.1 HACG01035440 CEK82305.1 KQ976423 KYM88749.1 GFAA01004163 JAT99271.1 ADTU01000888 AFFK01014253 AMQM01006515 AMQM01006516 KB097487 ESN96827.1 GDIP01235564 GDIP01038984 LRGB01000512 JAI87837.1 KZS18695.1 GDIP01199668 JAJ23734.1 HACG01035425 CEK82290.1 GDIQ01011400 JAN83337.1 GDIP01058789 JAM44926.1 GDIQ01138724 JAL13002.1 GEDV01004491 JAP84066.1 GFJQ02005092 JAW01878.1 GL732530 EFX86231.1 HACG01035419 CEK82284.1 KQ421523 KOF77227.1 GFPF01006413 MAA17559.1 GFPF01006415 MAA17561.1 HACG01035442 CEK82307.1 HACG01035439 CEK82304.1 APGK01045628 KB741037 ENN74658.1 BX510945
Proteomes
Pfam
PF00439 Bromodomain
Interpro
SUPFAM
SSF47370
SSF47370
Gene 3D
ProteinModelPortal
H9IWN4
A0A2A4K5G6
A0A212F487
S4PGH2
A0A1E1WLI2
A0A194Q4Z9
+ More
V5I6E6 E0VFW5 A0A067R8G4 A0A3S3PA68 A0A0P5HU95 A0A0P5HXK0 A0A0B7AN89 T1KG49 A0A026W9H9 A0A3L8DYM5 A0A3S1AXJ8 A0A210QL55 A0A3R7M2X0 A0A0B7AQV6 A0A0B7AQU7 A0A195BRB2 A0A1E1XJS8 A0A158NZL7 T1IIC5 T1FUX7 A0A0P4XX32 A0A0P5AH37 A0A0B7AQT4 A0A0P6J8B5 A0A0P5YIK9 A0A0P5PH60 A0A131Z0S7 A0A1Z5L3V3 E9G2D3 A0A0B7AMM0 A0A0L8GJP8 A0A224YP87 A0A224YSJ0 A0A0B7ANX6 A0A0B7AMP1 A0A1W4XC19 N6TAA1 E7F3M6 W5LHN2
V5I6E6 E0VFW5 A0A067R8G4 A0A3S3PA68 A0A0P5HU95 A0A0P5HXK0 A0A0B7AN89 T1KG49 A0A026W9H9 A0A3L8DYM5 A0A3S1AXJ8 A0A210QL55 A0A3R7M2X0 A0A0B7AQV6 A0A0B7AQU7 A0A195BRB2 A0A1E1XJS8 A0A158NZL7 T1IIC5 T1FUX7 A0A0P4XX32 A0A0P5AH37 A0A0B7AQT4 A0A0P6J8B5 A0A0P5YIK9 A0A0P5PH60 A0A131Z0S7 A0A1Z5L3V3 E9G2D3 A0A0B7AMM0 A0A0L8GJP8 A0A224YP87 A0A224YSJ0 A0A0B7ANX6 A0A0B7AMP1 A0A1W4XC19 N6TAA1 E7F3M6 W5LHN2
PDB
5C4Q
E-value=5.56402e-14,
Score=184
Ontologies
GO
PANTHER
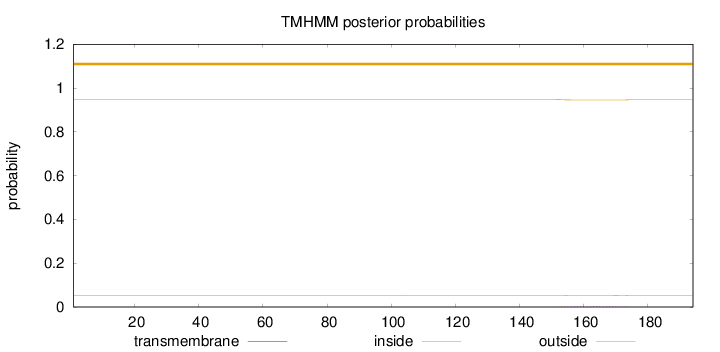
Topology
Length:
194
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05338
Exp number, first 60 AAs:
2e-05
Total prob of N-in:
0.05195
outside
1 - 194
Population Genetic Test Statistics
Pi
305.0787
Theta
151.815202
Tajima's D
3.699777
CLR
2.187401
CSRT
0.996350182490875
Interpretation
Uncertain
