Gene
KWMTBOMO06173
Pre Gene Modal
BGIBMGA001668
Annotation
Erg28-domain_containing_protein_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.463
Sequence
CDS
ATGCAGAACAAAGTTTTATACGCTTTCCGGGGATGGATAGCGTTCGTCGCGTTCATGGATTTGGGGACGGCCGGTCGATCGTATATCGAAGGAAGATCATTTCTCAATAACAATGGAGAGACTGAGCACTTAGACGGTGACTTCACGATATCGAGAATATTAGGAATGTATTCAGTTCTAAAAGCTCTAGCGCTCATACATTGCACCTTATACATACATTACAGACCTGTCGTCTCAATGGGTTACTGGTCGTTGATTCTAACGATAATATTGTATTTCACTGAAGCATTTTACTTTAGATCAACGAATCTAAATTTCTATGTGGTGTTTCCGTGCGTTTTGAACATTATCACACTTCTGGGTCTCGTGTACTTGCCGAACAAGCTGAGACTGTTTGGAACTGTACCCGGCACATCGGGCATGGACAGGGAGGTCGAAGACGAAAACGTTCAAATATTAAGACAAATGGGCAATTTCCGACGCAGAAAAACTGGCAACACTAAGAACAAACACGTCTAG
Protein
MQNKVLYAFRGWIAFVAFMDLGTAGRSYIEGRSFLNNNGETEHLDGDFTISRILGMYSVLKALALIHCTLYIHYRPVVSMGYWSLILTIILYFTEAFYFRSTNLNFYVVFPCVLNIITLLGLVYLPNKLRLFGTVPGTSGMDREVEDENVQILRQMGNFRRRKTGNTKNKHV
Summary
Uniprot
Q1HPH8
A0A0N1I8K8
A0A3S2NSN4
A0A2H1VJV7
A0A2A4JAU9
A0A0L0CJH0
+ More
W8AJN3 A0A182XPE8 A0A0K8USR4 B4NFT0 W5JPJ2 A0A182FP09 A0A0A1X368 A0A336MSD9 A0A182RE96 A0A182NE04 A0A3B0KHB5 A0A3B0KAG3 A0A1A9WNL3 B4JUD9 A0A1A9V8T4 A0A1B0A7T8 B4LWK5 A0A1B0CIX2 I5AN87 B3LZU7 B4GZF5 B4K8W3 A0A1W4VA45 A0A182GJI6 A0A0M3QXZ0 B3P326 B4R1S0 B4II73 Q9VDI6 A0A1I8NQV6 Q16NG9 B0WS43 B4PPS9 A0A1B0DP51 D6WKL0 A0A1L8D9B7 Q7Q7Q6 A0A1Y1KII4 A0A067RS16 A0A1Y1KQV5 A0A1B0FNS3 A0A1B6F7X7 A0A1J1HJE1 A0A1B6LZQ4 A0A1B6DEZ1 A0A1W4W4I5 A0A2R7VVT6 A0A1B6I1M2 R4FM59 A0A224XYK1 A0A1S4EQS3 A0A0A9WBI2 N6TDM6 J3JXK7 T1GJE3 A0A1Y1KKX0 Q16GU1 X1WKU0 E0VTQ1 A0A1D2MKD6 E9GL24 A0A0P5LX04 A0A0P5FAR4 A0A131Y2Q1 A0A0K8R4D8 A0A2G8LQF5 A0A1S3JPJ8 A0A1E1WWV6 C3XZD4 A0A3Q1H423 A0A3Q3VSP2 M3XH57 A0A2I4BK33 A0A3B4B4A3 A0A3B5KIH6
W8AJN3 A0A182XPE8 A0A0K8USR4 B4NFT0 W5JPJ2 A0A182FP09 A0A0A1X368 A0A336MSD9 A0A182RE96 A0A182NE04 A0A3B0KHB5 A0A3B0KAG3 A0A1A9WNL3 B4JUD9 A0A1A9V8T4 A0A1B0A7T8 B4LWK5 A0A1B0CIX2 I5AN87 B3LZU7 B4GZF5 B4K8W3 A0A1W4VA45 A0A182GJI6 A0A0M3QXZ0 B3P326 B4R1S0 B4II73 Q9VDI6 A0A1I8NQV6 Q16NG9 B0WS43 B4PPS9 A0A1B0DP51 D6WKL0 A0A1L8D9B7 Q7Q7Q6 A0A1Y1KII4 A0A067RS16 A0A1Y1KQV5 A0A1B0FNS3 A0A1B6F7X7 A0A1J1HJE1 A0A1B6LZQ4 A0A1B6DEZ1 A0A1W4W4I5 A0A2R7VVT6 A0A1B6I1M2 R4FM59 A0A224XYK1 A0A1S4EQS3 A0A0A9WBI2 N6TDM6 J3JXK7 T1GJE3 A0A1Y1KKX0 Q16GU1 X1WKU0 E0VTQ1 A0A1D2MKD6 E9GL24 A0A0P5LX04 A0A0P5FAR4 A0A131Y2Q1 A0A0K8R4D8 A0A2G8LQF5 A0A1S3JPJ8 A0A1E1WWV6 C3XZD4 A0A3Q1H423 A0A3Q3VSP2 M3XH57 A0A2I4BK33 A0A3B4B4A3 A0A3B5KIH6
Pubmed
26354079
26108605
24495485
17994087
20920257
23761445
+ More
25830018 15632085 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17510324 17550304 18362917 19820115 12364791 14747013 17210077 28004739 24845553 25401762 23537049 22516182 20566863 27289101 21292972 29652888 29023486 28503490 18563158 9215903 25463417 21551351
25830018 15632085 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17510324 17550304 18362917 19820115 12364791 14747013 17210077 28004739 24845553 25401762 23537049 22516182 20566863 27289101 21292972 29652888 29023486 28503490 18563158 9215903 25463417 21551351
EMBL
DQ443424
ABF51513.1
KQ460736
KPJ12605.1
RSAL01000101
RVE47520.1
+ More
ODYU01002956 SOQ41110.1 NWSH01002289 PCG68664.1 JRES01000310 KNC32386.1 GAMC01017755 JAB88800.1 GDHF01033516 GDHF01027491 GDHF01022751 JAI18798.1 JAI24823.1 JAI29563.1 CH964251 EDW83147.2 ADMH02000728 ETN65228.1 GBXI01008715 JAD05577.1 UFQS01001617 UFQT01001617 UFQT01001665 SSX11620.1 SSX31187.1 SSX31361.1 OUUW01000007 SPP83078.1 SPP83079.1 CH916374 EDV91109.1 CH940650 EDW67670.2 AJWK01013857 CM000070 EIM52422.2 CH902617 EDV43091.2 CH479198 EDW28173.1 CH933806 EDW16560.2 JXUM01068234 JXUM01068235 KQ562489 KXJ75796.1 CP012526 ALC46709.1 CH954181 EDV48340.1 CM000364 EDX12179.1 CH480842 EDW49617.1 AE014297 AY069274 AY071390 AAF55807.2 AAL39419.1 AAL49012.1 CH477824 EAT35894.1 DS232064 EDS33662.1 CM000160 EDW96179.2 AJVK01018085 AJVK01018086 KQ971342 EFA03564.2 GFDF01011154 JAV02930.1 AAAB01008952 EAA10621.4 EGK97246.1 GEZM01082822 JAV61259.1 KK852460 KDR23515.1 GEZM01082824 JAV61257.1 CCAG010016865 GECZ01031512 GECZ01023441 GECZ01003470 JAS38257.1 JAS46328.1 JAS66299.1 CVRI01000006 CRK88168.1 GEBQ01010810 JAT29167.1 GEDC01013096 JAS24202.1 KK854039 PTY10045.1 GECU01026894 JAS80812.1 ACPB03000063 GAHY01001849 JAA75661.1 GFTR01002746 JAW13680.1 GBHO01039721 GBRD01009379 JAG03883.1 JAG56445.1 APGK01041759 KB740997 KB632100 ENN75848.1 ERL88843.1 BT127976 AEE62938.1 CAQQ02086848 CAQQ02086849 GEZM01082823 JAV61258.1 CH478235 EAT33455.1 ABLF02030155 DS235769 EEB16757.1 LJIJ01000963 ODM93526.1 GL732550 EFX79779.1 GDIQ01188332 GDIP01054576 LRGB01002190 JAK63393.1 JAM49139.1 KZS08578.1 GDIQ01258177 JAJ93547.1 GEFM01002639 GEGO01005301 JAP73157.1 JAR90103.1 GADI01008070 JAA65738.1 MRZV01000011 PIK62489.1 GFAC01007675 JAT91513.1 GG666475 EEN66698.1 AFYH01003162 AFYH01003163
ODYU01002956 SOQ41110.1 NWSH01002289 PCG68664.1 JRES01000310 KNC32386.1 GAMC01017755 JAB88800.1 GDHF01033516 GDHF01027491 GDHF01022751 JAI18798.1 JAI24823.1 JAI29563.1 CH964251 EDW83147.2 ADMH02000728 ETN65228.1 GBXI01008715 JAD05577.1 UFQS01001617 UFQT01001617 UFQT01001665 SSX11620.1 SSX31187.1 SSX31361.1 OUUW01000007 SPP83078.1 SPP83079.1 CH916374 EDV91109.1 CH940650 EDW67670.2 AJWK01013857 CM000070 EIM52422.2 CH902617 EDV43091.2 CH479198 EDW28173.1 CH933806 EDW16560.2 JXUM01068234 JXUM01068235 KQ562489 KXJ75796.1 CP012526 ALC46709.1 CH954181 EDV48340.1 CM000364 EDX12179.1 CH480842 EDW49617.1 AE014297 AY069274 AY071390 AAF55807.2 AAL39419.1 AAL49012.1 CH477824 EAT35894.1 DS232064 EDS33662.1 CM000160 EDW96179.2 AJVK01018085 AJVK01018086 KQ971342 EFA03564.2 GFDF01011154 JAV02930.1 AAAB01008952 EAA10621.4 EGK97246.1 GEZM01082822 JAV61259.1 KK852460 KDR23515.1 GEZM01082824 JAV61257.1 CCAG010016865 GECZ01031512 GECZ01023441 GECZ01003470 JAS38257.1 JAS46328.1 JAS66299.1 CVRI01000006 CRK88168.1 GEBQ01010810 JAT29167.1 GEDC01013096 JAS24202.1 KK854039 PTY10045.1 GECU01026894 JAS80812.1 ACPB03000063 GAHY01001849 JAA75661.1 GFTR01002746 JAW13680.1 GBHO01039721 GBRD01009379 JAG03883.1 JAG56445.1 APGK01041759 KB740997 KB632100 ENN75848.1 ERL88843.1 BT127976 AEE62938.1 CAQQ02086848 CAQQ02086849 GEZM01082823 JAV61258.1 CH478235 EAT33455.1 ABLF02030155 DS235769 EEB16757.1 LJIJ01000963 ODM93526.1 GL732550 EFX79779.1 GDIQ01188332 GDIP01054576 LRGB01002190 JAK63393.1 JAM49139.1 KZS08578.1 GDIQ01258177 JAJ93547.1 GEFM01002639 GEGO01005301 JAP73157.1 JAR90103.1 GADI01008070 JAA65738.1 MRZV01000011 PIK62489.1 GFAC01007675 JAT91513.1 GG666475 EEN66698.1 AFYH01003162 AFYH01003163
Proteomes
UP000053240
UP000283053
UP000218220
UP000037069
UP000076407
UP000007798
+ More
UP000000673 UP000069272 UP000075900 UP000075884 UP000268350 UP000091820 UP000001070 UP000078200 UP000092445 UP000008792 UP000092461 UP000001819 UP000007801 UP000008744 UP000009192 UP000192221 UP000069940 UP000249989 UP000092553 UP000008711 UP000000304 UP000001292 UP000000803 UP000095300 UP000008820 UP000002320 UP000002282 UP000092462 UP000007266 UP000007062 UP000027135 UP000092444 UP000183832 UP000192223 UP000015103 UP000079169 UP000019118 UP000030742 UP000015102 UP000007819 UP000009046 UP000094527 UP000000305 UP000076858 UP000230750 UP000085678 UP000001554 UP000265040 UP000261620 UP000008672 UP000192220 UP000261520 UP000005226
UP000000673 UP000069272 UP000075900 UP000075884 UP000268350 UP000091820 UP000001070 UP000078200 UP000092445 UP000008792 UP000092461 UP000001819 UP000007801 UP000008744 UP000009192 UP000192221 UP000069940 UP000249989 UP000092553 UP000008711 UP000000304 UP000001292 UP000000803 UP000095300 UP000008820 UP000002320 UP000002282 UP000092462 UP000007266 UP000007062 UP000027135 UP000092444 UP000183832 UP000192223 UP000015103 UP000079169 UP000019118 UP000030742 UP000015102 UP000007819 UP000009046 UP000094527 UP000000305 UP000076858 UP000230750 UP000085678 UP000001554 UP000265040 UP000261620 UP000008672 UP000192220 UP000261520 UP000005226
Pfam
PF03694 Erg28
Interpro
IPR005352
Erg28
ProteinModelPortal
Q1HPH8
A0A0N1I8K8
A0A3S2NSN4
A0A2H1VJV7
A0A2A4JAU9
A0A0L0CJH0
+ More
W8AJN3 A0A182XPE8 A0A0K8USR4 B4NFT0 W5JPJ2 A0A182FP09 A0A0A1X368 A0A336MSD9 A0A182RE96 A0A182NE04 A0A3B0KHB5 A0A3B0KAG3 A0A1A9WNL3 B4JUD9 A0A1A9V8T4 A0A1B0A7T8 B4LWK5 A0A1B0CIX2 I5AN87 B3LZU7 B4GZF5 B4K8W3 A0A1W4VA45 A0A182GJI6 A0A0M3QXZ0 B3P326 B4R1S0 B4II73 Q9VDI6 A0A1I8NQV6 Q16NG9 B0WS43 B4PPS9 A0A1B0DP51 D6WKL0 A0A1L8D9B7 Q7Q7Q6 A0A1Y1KII4 A0A067RS16 A0A1Y1KQV5 A0A1B0FNS3 A0A1B6F7X7 A0A1J1HJE1 A0A1B6LZQ4 A0A1B6DEZ1 A0A1W4W4I5 A0A2R7VVT6 A0A1B6I1M2 R4FM59 A0A224XYK1 A0A1S4EQS3 A0A0A9WBI2 N6TDM6 J3JXK7 T1GJE3 A0A1Y1KKX0 Q16GU1 X1WKU0 E0VTQ1 A0A1D2MKD6 E9GL24 A0A0P5LX04 A0A0P5FAR4 A0A131Y2Q1 A0A0K8R4D8 A0A2G8LQF5 A0A1S3JPJ8 A0A1E1WWV6 C3XZD4 A0A3Q1H423 A0A3Q3VSP2 M3XH57 A0A2I4BK33 A0A3B4B4A3 A0A3B5KIH6
W8AJN3 A0A182XPE8 A0A0K8USR4 B4NFT0 W5JPJ2 A0A182FP09 A0A0A1X368 A0A336MSD9 A0A182RE96 A0A182NE04 A0A3B0KHB5 A0A3B0KAG3 A0A1A9WNL3 B4JUD9 A0A1A9V8T4 A0A1B0A7T8 B4LWK5 A0A1B0CIX2 I5AN87 B3LZU7 B4GZF5 B4K8W3 A0A1W4VA45 A0A182GJI6 A0A0M3QXZ0 B3P326 B4R1S0 B4II73 Q9VDI6 A0A1I8NQV6 Q16NG9 B0WS43 B4PPS9 A0A1B0DP51 D6WKL0 A0A1L8D9B7 Q7Q7Q6 A0A1Y1KII4 A0A067RS16 A0A1Y1KQV5 A0A1B0FNS3 A0A1B6F7X7 A0A1J1HJE1 A0A1B6LZQ4 A0A1B6DEZ1 A0A1W4W4I5 A0A2R7VVT6 A0A1B6I1M2 R4FM59 A0A224XYK1 A0A1S4EQS3 A0A0A9WBI2 N6TDM6 J3JXK7 T1GJE3 A0A1Y1KKX0 Q16GU1 X1WKU0 E0VTQ1 A0A1D2MKD6 E9GL24 A0A0P5LX04 A0A0P5FAR4 A0A131Y2Q1 A0A0K8R4D8 A0A2G8LQF5 A0A1S3JPJ8 A0A1E1WWV6 C3XZD4 A0A3Q1H423 A0A3Q3VSP2 M3XH57 A0A2I4BK33 A0A3B4B4A3 A0A3B5KIH6
Ontologies
GO
PANTHER
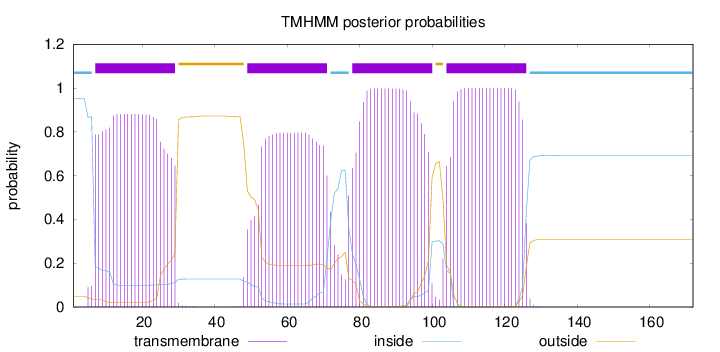
Topology
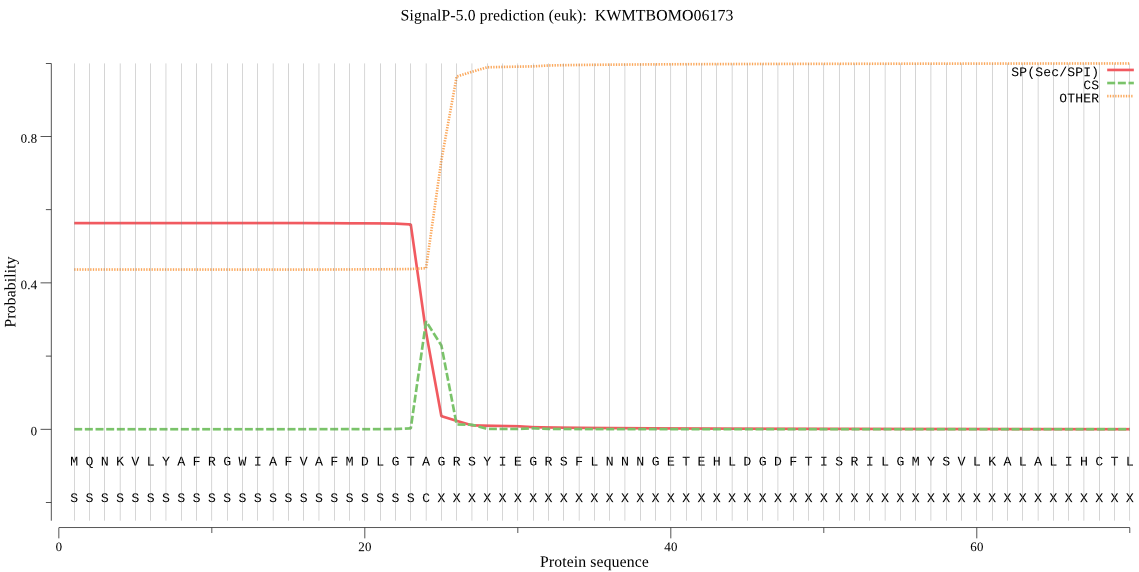
SignalP
Position: 1 - 24,
Likelihood: 0.563247
Length:
172
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
79.24778
Exp number, first 60 AAs:
27.16591
Total prob of N-in:
0.95154
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 48
TMhelix
49 - 71
inside
72 - 77
TMhelix
78 - 100
outside
101 - 103
TMhelix
104 - 126
inside
127 - 172
Population Genetic Test Statistics
Pi
251.806047
Theta
184.622404
Tajima's D
1.472783
CLR
0.074815
CSRT
0.779911004449777
Interpretation
Uncertain
