Pre Gene Modal
BGIBMGA001767
Annotation
calmodulin-like_protein_4_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.112 Mitochondrial Reliability : 1.136
Sequence
CDS
ATGTCGTTCGCGGATTTCCTTGAAGTTATGCACATACATTCTCGTGCCGAAAATCTTCCGTCAGAGGTAGTAAACGCGTTTAAGGCCGGAGATCCGAACAAGACCGGAGTGATATCGGCTAAGCAACTGCGGACGCTTCTGCAGAAATGGGGAGAAGGGCTCTCAGCCAGAGAAGTAGACAACATTTTCCGCGAAGCGAATGTTTCCAACAGTGCTACGGTGCGCTACGAAGACTTCGTTAAAATCGCCTGCGCCCCAGTTCCGGACTATTATTAA
Protein
MSFADFLEVMHIHSRAENLPSEVVNAFKAGDPNKTGVISAKQLRTLLQKWGEGLSAREVDNIFREANVSNSATVRYEDFVKIACAPVPDYY
Summary
Uniprot
H9IWY4
A0A3S2LM22
A0A2W1BAL7
A0A2A4J9A7
I4DKR4
A0A2W1B515
+ More
A0A194Q4F6 A0A0N1IF97 S4PAJ0 A0A1Y1LS43 U5ENA1 A0A0L0CJG8 D6WML6 A0A0K8TRA9 A0A1B0CRI0 A0A1B0CZ74 A0A0Q9WGE9 B4JUE2 A0A1I8NN02 A0A1L8EE69 A0A0K8UKM6 B4K8W5 A0A1W4V9U2 B4R1R6 B4PPT3 B3M342 B3P322 Q9VDI3 B4NFS6 B4II69 A0A034WVF2 A0A0A1WTM9 A0A0K8W7E5 A0A1A9WNL8 A0A182RE97 W8CB87 A0A1I8MQQ1 A0A182YER9 A0A1Q3FLM7 A0A182T827 A0A1L8DND4 A0A0M5J502 A0A1L8DNB4 A0A182LNP9 A0A182XDR8 Q7Q7Q5 A0A182IDH7 A0A2M4APT6 A0A182NE03 A0A2M4AQ89 A0A3B0K1R2 B0WS44 A0A182QDG0 A0A1W4WGY7 A0A182VPS3 J3JW20 A0A182J752 A0A1B0FNR6 A0A1B0A7U1 A0A1A9V8S8 N6T323 Q16NH0 A0A023EGS9 T1GRL9 A0A1B0BNT8 A0A182UP82 T1DQ39 A0A2M3ZI77 A0A1Y9G9J6 A0A2M4C2E5 W5JLH5 A0A182M5A2 A0A1A9XM88 Q299D4 B4G519 A0A2P8Z6R7 R4V3T9 A0A2J7Q4K8 A0A1B6H8S4 A0A067R676 A0A1B6MLF1 A0A1B6EPI7 A0A336MMW7 A0A0A9ZE84 A0A1S3DM60 A0A336MZ73 A0A1S4EFG6 T1ICB4 T1DDR2 A0A171AUV7 A0A151WKN1 A0A088AF37 A0A158NZC6 A0A151IKB5 A0A026WAK8 A0A069DNT4 K7IMH9 A0A2R7WCS3 F4W9W3 E0VBH9
A0A194Q4F6 A0A0N1IF97 S4PAJ0 A0A1Y1LS43 U5ENA1 A0A0L0CJG8 D6WML6 A0A0K8TRA9 A0A1B0CRI0 A0A1B0CZ74 A0A0Q9WGE9 B4JUE2 A0A1I8NN02 A0A1L8EE69 A0A0K8UKM6 B4K8W5 A0A1W4V9U2 B4R1R6 B4PPT3 B3M342 B3P322 Q9VDI3 B4NFS6 B4II69 A0A034WVF2 A0A0A1WTM9 A0A0K8W7E5 A0A1A9WNL8 A0A182RE97 W8CB87 A0A1I8MQQ1 A0A182YER9 A0A1Q3FLM7 A0A182T827 A0A1L8DND4 A0A0M5J502 A0A1L8DNB4 A0A182LNP9 A0A182XDR8 Q7Q7Q5 A0A182IDH7 A0A2M4APT6 A0A182NE03 A0A2M4AQ89 A0A3B0K1R2 B0WS44 A0A182QDG0 A0A1W4WGY7 A0A182VPS3 J3JW20 A0A182J752 A0A1B0FNR6 A0A1B0A7U1 A0A1A9V8S8 N6T323 Q16NH0 A0A023EGS9 T1GRL9 A0A1B0BNT8 A0A182UP82 T1DQ39 A0A2M3ZI77 A0A1Y9G9J6 A0A2M4C2E5 W5JLH5 A0A182M5A2 A0A1A9XM88 Q299D4 B4G519 A0A2P8Z6R7 R4V3T9 A0A2J7Q4K8 A0A1B6H8S4 A0A067R676 A0A1B6MLF1 A0A1B6EPI7 A0A336MMW7 A0A0A9ZE84 A0A1S3DM60 A0A336MZ73 A0A1S4EFG6 T1ICB4 T1DDR2 A0A171AUV7 A0A151WKN1 A0A088AF37 A0A158NZC6 A0A151IKB5 A0A026WAK8 A0A069DNT4 K7IMH9 A0A2R7WCS3 F4W9W3 E0VBH9
Pubmed
19121390
28756777
22651552
26354079
23622113
28004739
+ More
26108605 18362917 19820115 26369729 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 25830018 24495485 25315136 25244985 20966253 12364791 14747013 17210077 22516182 23537049 17510324 24945155 26483478 20920257 23761445 15632085 29403074 24845553 25401762 26823975 24330624 21347285 24508170 26334808 20075255 21719571 20566863
26108605 18362917 19820115 26369729 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 25830018 24495485 25315136 25244985 20966253 12364791 14747013 17210077 22516182 23537049 17510324 24945155 26483478 20920257 23761445 15632085 29403074 24845553 25401762 26823975 24330624 21347285 24508170 26334808 20075255 21719571 20566863
EMBL
BABH01021133
BABH01021134
RSAL01003921
RSAL01000101
RVE40215.1
RVE47521.1
+ More
KZ150592 PZC70477.1 NWSH01002289 PCG68665.1 AK401882 BAM18504.1 KZ150548 PZC70561.1 KQ459463 KPJ00427.1 KQ460736 KPJ12604.1 GAIX01006357 JAA86203.1 GEZM01051214 GEZM01051213 GEZM01051211 JAV75090.1 GANO01000679 JAB59192.1 JRES01000310 KNC32380.1 KQ971343 EFA04290.1 GDAI01000696 JAI16907.1 AJWK01024813 AJVK01020669 CH940650 KRF83386.1 CH916374 EDV91112.1 GFDG01001833 JAV16966.1 GDHF01025216 JAI27098.1 CH933806 EDW16562.2 CM000364 EDX12175.1 CM000160 EDW96183.2 CH902617 EDV42442.2 CH954181 EDV48336.1 AE014297 AY071049 KX532102 AAF55810.2 AAL48671.1 ANY27912.1 CH964251 EDW83143.2 CH480842 EDW49613.1 GAKP01000338 JAC58614.1 GBXI01012447 GBXI01010227 JAD01845.1 JAD04065.1 GDHF01005544 JAI46770.1 GAMC01000674 JAC05882.1 GFDL01006613 JAV28432.1 GFDF01006210 JAV07874.1 CP012526 ALC46711.1 GFDF01006230 JAV07854.1 AAAB01008952 EAA10553.4 APCN01005980 GGFK01009480 MBW42801.1 GGFK01009612 MBW42933.1 OUUW01000013 SPP88174.1 DS232064 EDS33663.1 AXCN02002212 BT127438 KB632305 AEE62400.1 ERL91848.1 CCAG010016865 APGK01054266 KB741248 ENN71953.1 CH477824 EAT35893.1 JXUM01068236 GAPW01005562 KQ562489 JAC08036.1 KXJ75798.1 CAQQ02100334 CAQQ02100335 JXJN01017603 GAMD01002794 JAA98796.1 GGFM01007516 MBW28267.1 GGFJ01010349 MBW59490.1 ADMH02000728 GGFL01004383 ETN65227.1 MBW68561.1 AXCM01000238 CM000070 EAL27769.3 CH479179 EDW24685.1 PYGN01000171 PSN52188.1 KC571974 AGM32473.1 NEVH01018378 PNF23506.1 GECU01036646 JAS71060.1 KK852672 KDR18760.1 GEBQ01003282 JAT36695.1 GECZ01029933 JAS39836.1 UFQT01001665 SSX31360.1 GBHO01009685 GBHO01001956 GBRD01009247 GDHC01001329 JAG33919.1 JAG41648.1 JAG56574.1 JAQ17300.1 UFQS01001617 UFQT01001617 SSX11621.1 SSX31188.1 ACPB03019765 GALA01001347 JAA93505.1 GEMB01000628 JAS02505.1 KQ983012 KYQ48367.1 ADTU01000884 KQ977234 KYN04698.1 KK107321 EZA52691.1 GBGD01003404 JAC85485.1 KK854633 PTY17484.1 GL888033 EGI69099.1 DS235030 EEB10735.1
KZ150592 PZC70477.1 NWSH01002289 PCG68665.1 AK401882 BAM18504.1 KZ150548 PZC70561.1 KQ459463 KPJ00427.1 KQ460736 KPJ12604.1 GAIX01006357 JAA86203.1 GEZM01051214 GEZM01051213 GEZM01051211 JAV75090.1 GANO01000679 JAB59192.1 JRES01000310 KNC32380.1 KQ971343 EFA04290.1 GDAI01000696 JAI16907.1 AJWK01024813 AJVK01020669 CH940650 KRF83386.1 CH916374 EDV91112.1 GFDG01001833 JAV16966.1 GDHF01025216 JAI27098.1 CH933806 EDW16562.2 CM000364 EDX12175.1 CM000160 EDW96183.2 CH902617 EDV42442.2 CH954181 EDV48336.1 AE014297 AY071049 KX532102 AAF55810.2 AAL48671.1 ANY27912.1 CH964251 EDW83143.2 CH480842 EDW49613.1 GAKP01000338 JAC58614.1 GBXI01012447 GBXI01010227 JAD01845.1 JAD04065.1 GDHF01005544 JAI46770.1 GAMC01000674 JAC05882.1 GFDL01006613 JAV28432.1 GFDF01006210 JAV07874.1 CP012526 ALC46711.1 GFDF01006230 JAV07854.1 AAAB01008952 EAA10553.4 APCN01005980 GGFK01009480 MBW42801.1 GGFK01009612 MBW42933.1 OUUW01000013 SPP88174.1 DS232064 EDS33663.1 AXCN02002212 BT127438 KB632305 AEE62400.1 ERL91848.1 CCAG010016865 APGK01054266 KB741248 ENN71953.1 CH477824 EAT35893.1 JXUM01068236 GAPW01005562 KQ562489 JAC08036.1 KXJ75798.1 CAQQ02100334 CAQQ02100335 JXJN01017603 GAMD01002794 JAA98796.1 GGFM01007516 MBW28267.1 GGFJ01010349 MBW59490.1 ADMH02000728 GGFL01004383 ETN65227.1 MBW68561.1 AXCM01000238 CM000070 EAL27769.3 CH479179 EDW24685.1 PYGN01000171 PSN52188.1 KC571974 AGM32473.1 NEVH01018378 PNF23506.1 GECU01036646 JAS71060.1 KK852672 KDR18760.1 GEBQ01003282 JAT36695.1 GECZ01029933 JAS39836.1 UFQT01001665 SSX31360.1 GBHO01009685 GBHO01001956 GBRD01009247 GDHC01001329 JAG33919.1 JAG41648.1 JAG56574.1 JAQ17300.1 UFQS01001617 UFQT01001617 SSX11621.1 SSX31188.1 ACPB03019765 GALA01001347 JAA93505.1 GEMB01000628 JAS02505.1 KQ983012 KYQ48367.1 ADTU01000884 KQ977234 KYN04698.1 KK107321 EZA52691.1 GBGD01003404 JAC85485.1 KK854633 PTY17484.1 GL888033 EGI69099.1 DS235030 EEB10735.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000037069
+ More
UP000007266 UP000092461 UP000092462 UP000008792 UP000001070 UP000095300 UP000009192 UP000192221 UP000000304 UP000002282 UP000007801 UP000008711 UP000000803 UP000007798 UP000001292 UP000091820 UP000075900 UP000095301 UP000076408 UP000075901 UP000092553 UP000075882 UP000076407 UP000007062 UP000075840 UP000075884 UP000268350 UP000002320 UP000075886 UP000192223 UP000075920 UP000030742 UP000075880 UP000092444 UP000092445 UP000078200 UP000019118 UP000008820 UP000069940 UP000249989 UP000015102 UP000092460 UP000075903 UP000069272 UP000000673 UP000075883 UP000092443 UP000001819 UP000008744 UP000245037 UP000235965 UP000027135 UP000079169 UP000015103 UP000075809 UP000005203 UP000005205 UP000078542 UP000053097 UP000002358 UP000007755 UP000009046
UP000007266 UP000092461 UP000092462 UP000008792 UP000001070 UP000095300 UP000009192 UP000192221 UP000000304 UP000002282 UP000007801 UP000008711 UP000000803 UP000007798 UP000001292 UP000091820 UP000075900 UP000095301 UP000076408 UP000075901 UP000092553 UP000075882 UP000076407 UP000007062 UP000075840 UP000075884 UP000268350 UP000002320 UP000075886 UP000192223 UP000075920 UP000030742 UP000075880 UP000092444 UP000092445 UP000078200 UP000019118 UP000008820 UP000069940 UP000249989 UP000015102 UP000092460 UP000075903 UP000069272 UP000000673 UP000075883 UP000092443 UP000001819 UP000008744 UP000245037 UP000235965 UP000027135 UP000079169 UP000015103 UP000075809 UP000005203 UP000005205 UP000078542 UP000053097 UP000002358 UP000007755 UP000009046
Interpro
Gene 3D
CDD
ProteinModelPortal
H9IWY4
A0A3S2LM22
A0A2W1BAL7
A0A2A4J9A7
I4DKR4
A0A2W1B515
+ More
A0A194Q4F6 A0A0N1IF97 S4PAJ0 A0A1Y1LS43 U5ENA1 A0A0L0CJG8 D6WML6 A0A0K8TRA9 A0A1B0CRI0 A0A1B0CZ74 A0A0Q9WGE9 B4JUE2 A0A1I8NN02 A0A1L8EE69 A0A0K8UKM6 B4K8W5 A0A1W4V9U2 B4R1R6 B4PPT3 B3M342 B3P322 Q9VDI3 B4NFS6 B4II69 A0A034WVF2 A0A0A1WTM9 A0A0K8W7E5 A0A1A9WNL8 A0A182RE97 W8CB87 A0A1I8MQQ1 A0A182YER9 A0A1Q3FLM7 A0A182T827 A0A1L8DND4 A0A0M5J502 A0A1L8DNB4 A0A182LNP9 A0A182XDR8 Q7Q7Q5 A0A182IDH7 A0A2M4APT6 A0A182NE03 A0A2M4AQ89 A0A3B0K1R2 B0WS44 A0A182QDG0 A0A1W4WGY7 A0A182VPS3 J3JW20 A0A182J752 A0A1B0FNR6 A0A1B0A7U1 A0A1A9V8S8 N6T323 Q16NH0 A0A023EGS9 T1GRL9 A0A1B0BNT8 A0A182UP82 T1DQ39 A0A2M3ZI77 A0A1Y9G9J6 A0A2M4C2E5 W5JLH5 A0A182M5A2 A0A1A9XM88 Q299D4 B4G519 A0A2P8Z6R7 R4V3T9 A0A2J7Q4K8 A0A1B6H8S4 A0A067R676 A0A1B6MLF1 A0A1B6EPI7 A0A336MMW7 A0A0A9ZE84 A0A1S3DM60 A0A336MZ73 A0A1S4EFG6 T1ICB4 T1DDR2 A0A171AUV7 A0A151WKN1 A0A088AF37 A0A158NZC6 A0A151IKB5 A0A026WAK8 A0A069DNT4 K7IMH9 A0A2R7WCS3 F4W9W3 E0VBH9
A0A194Q4F6 A0A0N1IF97 S4PAJ0 A0A1Y1LS43 U5ENA1 A0A0L0CJG8 D6WML6 A0A0K8TRA9 A0A1B0CRI0 A0A1B0CZ74 A0A0Q9WGE9 B4JUE2 A0A1I8NN02 A0A1L8EE69 A0A0K8UKM6 B4K8W5 A0A1W4V9U2 B4R1R6 B4PPT3 B3M342 B3P322 Q9VDI3 B4NFS6 B4II69 A0A034WVF2 A0A0A1WTM9 A0A0K8W7E5 A0A1A9WNL8 A0A182RE97 W8CB87 A0A1I8MQQ1 A0A182YER9 A0A1Q3FLM7 A0A182T827 A0A1L8DND4 A0A0M5J502 A0A1L8DNB4 A0A182LNP9 A0A182XDR8 Q7Q7Q5 A0A182IDH7 A0A2M4APT6 A0A182NE03 A0A2M4AQ89 A0A3B0K1R2 B0WS44 A0A182QDG0 A0A1W4WGY7 A0A182VPS3 J3JW20 A0A182J752 A0A1B0FNR6 A0A1B0A7U1 A0A1A9V8S8 N6T323 Q16NH0 A0A023EGS9 T1GRL9 A0A1B0BNT8 A0A182UP82 T1DQ39 A0A2M3ZI77 A0A1Y9G9J6 A0A2M4C2E5 W5JLH5 A0A182M5A2 A0A1A9XM88 Q299D4 B4G519 A0A2P8Z6R7 R4V3T9 A0A2J7Q4K8 A0A1B6H8S4 A0A067R676 A0A1B6MLF1 A0A1B6EPI7 A0A336MMW7 A0A0A9ZE84 A0A1S3DM60 A0A336MZ73 A0A1S4EFG6 T1ICB4 T1DDR2 A0A171AUV7 A0A151WKN1 A0A088AF37 A0A158NZC6 A0A151IKB5 A0A026WAK8 A0A069DNT4 K7IMH9 A0A2R7WCS3 F4W9W3 E0VBH9
PDB
5A2H
E-value=1.25031e-09,
Score=144
Ontologies
GO
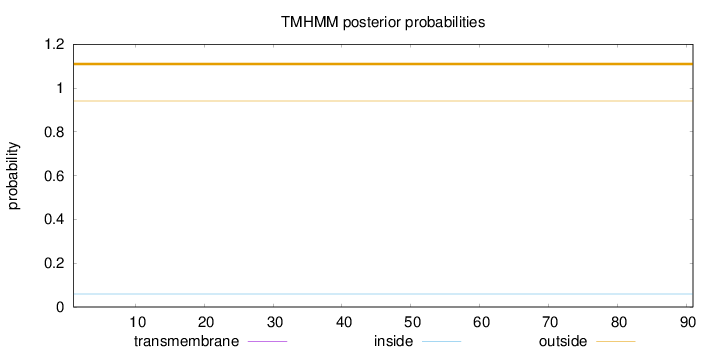
Topology
Length:
91
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05972
outside
1 - 91
Population Genetic Test Statistics
Pi
284.009085
Theta
200.231986
Tajima's D
1.267001
CLR
1.357185
CSRT
0.730463476826159
Interpretation
Uncertain
