Gene
KWMTBOMO06170
Pre Gene Modal
BGIBMGA001766
Annotation
Phosphatidylserine_decarboxylase_[Operophtera_brumata]
Full name
Phosphatidylserine decarboxylase proenzyme, mitochondrial
Location in the cell
Mitochondrial Reliability : 1.407 PlasmaMembrane Reliability : 1.779
Sequence
CDS
ATGTTCCCCTTCCGGGTGACCAGTCGATTGTGGGGGAAAATGGCGGCCTGCGAAATCCCGGTGTCCCTCCGCAGCTTCGTCTACGGCACGTACATAAGAATGTTCAGCGTGAACCTAAATGATGCAGCAGTCACCGACCTGAAATACTACAAAAGCTTATCAGCGTTCTTCACCAGGCCGCTCAGGGATGGCGCGAGATACATATCAGCAGCGCCCTGCGTATCGCCCTGTGACGGAGTAGTCTTGAACTGTGGCCCAGCTGATACCGATAAGATTGAACAGGTAAAAGGTGTGACATACAGCCTTGAGGAATTCCTCGGTGAAAACAAATGGCTAAAAAGGAAAGATGAGAGCTATTACAACTCCTTACTAAAGAATAAAGAGAATATATTACATCAGTGCATAATATATTTAGCGCCCGGGGATTATCACCGCTTTCATGCTCCTTGTGATTGGACTGCTACATTCAGGAGACATTTCTCAGGGAAATTGCTCTCTGTAAATCCTTGGTTGGCGAAACTAATACCTGGATTGTTCACTGTCAACGAAAGAGCTGTGTACGTCGGAGAATGGGAGTATGGCTTCTTCAGCATGACAGCCGTCGGAGCGACAAATGTGGGATCTATAGAAATATTCAAAGATCCAGAATTAAGAACGAACACTAAAGGTAAACGGAATCGTGTGAACGAACTCGAATTAGGGCAGGTATGCATGAGCAAAGGTGAACTCTTCGGTCAGTTCAATATGGGAAGTACTATCATACTGCTCTTTGAGGCCCCAAAGGACTTCAAATTTGATATGGCTGCTGGAGATAAAGTATTAGTTGGACAATCATTAACGAAAGTAACCAGACAAATGACGAGATGA
Protein
MFPFRVTSRLWGKMAACEIPVSLRSFVYGTYIRMFSVNLNDAAVTDLKYYKSLSAFFTRPLRDGARYISAAPCVSPCDGVVLNCGPADTDKIEQVKGVTYSLEEFLGENKWLKRKDESYYNSLLKNKENILHQCIIYLAPGDYHRFHAPCDWTATFRRHFSGKLLSVNPWLAKLIPGLFTVNERAVYVGEWEYGFFSMTAVGATNVGSIEIFKDPELRTNTKGKRNRVNELELGQVCMSKGELFGQFNMGSTIILLFEAPKDFKFDMAAGDKVLVGQSLTKVTRQMTR
Summary
Description
Catalyzes the formation of phosphatidylethanolamine (PtdEtn) from phosphatidylserine (PtdSer). Plays a central role in phospholipid metabolism and in the interorganelle trafficking of phosphatidylserine.
Catalytic Activity
1,2-diacyl-sn-glycero-3-phospho-L-serine + H(+) = a 1,2-diacyl-sn-glycero-3-phosphoethanolamine + CO2
Cofactor
pyruvate
Subunit
Heterodimer of a large membrane-associated beta subunit and a small pyruvoyl-containing alpha subunit.
Similarity
Belongs to the phosphatidylserine decarboxylase family. PSD-B subfamily. Eukaryotic type I sub-subfamily.
Feature
chain Phosphatidylserine decarboxylase proenzyme, mitochondrial
Uniprot
H9IWY3
A0A2A4JCW4
A0A2W1BHW7
A0A0L7L7K7
A0A2H1WC49
A0A212FE93
+ More
A0A0N0PC89 A0A194QAI4 A0A1B0G858 A0A0L0CLH3 B3M2S6 A0A067R9S4 A0A1Y1LCI2 A0A1A9XI27 A0A1B0BJP7 A0A0M4EP02 A0A1I8NB19 T1PGF3 J3JWY2 U4UMN9 A0A084W044 U5ES05 B4NBP0 A0A1A9VR18 A0A3B0K428 Q298L0 A0A1I8PGN0 B4G433 A0A023EQ94 A0A182J586 W8AZA7 A0A1Q3FWK8 A0A1Q3FW75 A0A1Q3FW18 A0A1Q3FVW1 B4K5A4 A0A034WFZ6 Q16TJ5 B4QSF4 B4PL28 A0A336LR80 A0A1A9X1B4 B4MBZ9 A0A1J1IQ26 A0A1W4V4T4 A0A1S4FPW9 K7IMK1 A0A2M3Z8T6 W5J6W7 A0A182PRT9 Q9VCE0 A0A0K8WII1 A0A182GM50 B4HGH0 B3P7A3 A0A232EWZ2 A0A182M5N0 A0A182QSB2 V5I9R0 A0A0A1WFB6 A0A182NQQ8 C6TPB6 A0A139WNM4 A0A2M4AT34 B4JIH6 Q7PX64 A0A182X9E5 A0A182RSB8 A0A182VHL7 A0A182TKY9 A0A087ZIK7 A0A182FS14 A0A182YHY5 A0A1Y9IXW8 A0A182HKD1 A0A182L8L6 A0A182JUS1 A0A1B0CIY5 A0A2J7R5S6 A0A0P6GF40 A0A0P5NGP9 A0A1L8DZ44 T1K3B0 A0A0P5MRD9 A0A0P5ZC79 A0A0P5JXN8 A0A0P5MJB2 A0A0P5XLG0 A0A0P5MBV6 A0A0P5TN97 A0A0P5M1H6 A0A0N8BYV3 A0A0P5DE76 A0A0N8CEY7 A0A0P5LPF9 A0A0N8EC00 A0A0N8C7Q2 A0A0P6DLD2 A0A1L8DY97 A0A1L8DY52
A0A0N0PC89 A0A194QAI4 A0A1B0G858 A0A0L0CLH3 B3M2S6 A0A067R9S4 A0A1Y1LCI2 A0A1A9XI27 A0A1B0BJP7 A0A0M4EP02 A0A1I8NB19 T1PGF3 J3JWY2 U4UMN9 A0A084W044 U5ES05 B4NBP0 A0A1A9VR18 A0A3B0K428 Q298L0 A0A1I8PGN0 B4G433 A0A023EQ94 A0A182J586 W8AZA7 A0A1Q3FWK8 A0A1Q3FW75 A0A1Q3FW18 A0A1Q3FVW1 B4K5A4 A0A034WFZ6 Q16TJ5 B4QSF4 B4PL28 A0A336LR80 A0A1A9X1B4 B4MBZ9 A0A1J1IQ26 A0A1W4V4T4 A0A1S4FPW9 K7IMK1 A0A2M3Z8T6 W5J6W7 A0A182PRT9 Q9VCE0 A0A0K8WII1 A0A182GM50 B4HGH0 B3P7A3 A0A232EWZ2 A0A182M5N0 A0A182QSB2 V5I9R0 A0A0A1WFB6 A0A182NQQ8 C6TPB6 A0A139WNM4 A0A2M4AT34 B4JIH6 Q7PX64 A0A182X9E5 A0A182RSB8 A0A182VHL7 A0A182TKY9 A0A087ZIK7 A0A182FS14 A0A182YHY5 A0A1Y9IXW8 A0A182HKD1 A0A182L8L6 A0A182JUS1 A0A1B0CIY5 A0A2J7R5S6 A0A0P6GF40 A0A0P5NGP9 A0A1L8DZ44 T1K3B0 A0A0P5MRD9 A0A0P5ZC79 A0A0P5JXN8 A0A0P5MJB2 A0A0P5XLG0 A0A0P5MBV6 A0A0P5TN97 A0A0P5M1H6 A0A0N8BYV3 A0A0P5DE76 A0A0N8CEY7 A0A0P5LPF9 A0A0N8EC00 A0A0N8C7Q2 A0A0P6DLD2 A0A1L8DY97 A0A1L8DY52
EC Number
4.1.1.65
Pubmed
19121390
28756777
26227816
22118469
26354079
26108605
+ More
17994087 24845553 28004739 25315136 22516182 23537049 24438588 15632085 24945155 24495485 25348373 17510324 17550304 20075255 20920257 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 28648823 25830018 18362917 19820115 12364791 14747013 17210077 25244985 20966253
17994087 24845553 28004739 25315136 22516182 23537049 24438588 15632085 24945155 24495485 25348373 17510324 17550304 20075255 20920257 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 28648823 25830018 18362917 19820115 12364791 14747013 17210077 25244985 20966253
EMBL
BABH01021133
NWSH01002066
PCG69262.1
KZ150350
PZC71263.1
JTDY01002484
+ More
KOB71324.1 ODYU01007549 SOQ50412.1 AGBW02008966 OWR52051.1 KQ460736 KPJ12601.1 KQ459463 KPJ00426.1 CCAG010000199 JRES01000321 KNC32294.1 CH902617 EDV42397.1 KK852832 KDR15313.1 GEZM01061243 JAV70608.1 JXJN01015477 CP012526 ALC47440.1 KA647220 AFP61849.1 APGK01039349 APGK01039350 BT127750 KB740970 AEE62712.1 ENN76643.1 KB632388 ERL94372.1 ATLV01019085 KE525262 KFB43588.1 GANO01003475 JAB56396.1 CH964232 EDW81204.1 OUUW01000007 SPP82730.1 CM000070 EAL27945.1 CH479179 EDW24444.1 GAPW01002363 JAC11235.1 GAMC01014978 JAB91577.1 GFDL01003242 JAV31803.1 GFDL01003240 JAV31805.1 GFDL01003300 JAV31745.1 GFDL01003306 JAV31739.1 CH933806 EDW15104.1 GAKP01004441 JAC54511.1 CH477648 EAT37827.1 CM000364 EDX14153.1 CM000160 EDW99013.1 UFQS01000083 UFQT01000083 SSW99069.1 SSX19451.1 CH940656 EDW58620.1 CVRI01000057 CRL02264.1 GGFM01004149 MBW24900.1 ADMH02002101 ETN59143.1 AE014297 AY121648 AAF56228.1 AAM51975.1 GDHF01001353 JAI50961.1 JXUM01073817 JXUM01073818 JXUM01073819 KQ562796 KXJ75113.1 CH480815 EDW43423.1 CH954182 EDV53923.1 NNAY01001807 OXU22866.1 AXCM01002705 AXCN02001502 GALX01002500 JAB65966.1 GBXI01017117 JAC97174.1 BT099602 ACU45755.1 KQ971311 KYB29477.1 GGFK01010561 MBW43882.1 CH916369 EDV93057.1 AAAB01008987 EAA01787.4 EGK97628.1 APCN01000826 AJWK01013903 NEVH01006990 PNF36186.1 GDIQ01033973 JAN60764.1 GDIQ01153078 JAK98647.1 GFDF01002532 JAV11552.1 CAEY01001378 GDIQ01153079 JAK98646.1 GDIP01058605 JAM45110.1 GDIQ01218256 JAK33469.1 GDIQ01155777 JAK95948.1 GDIP01070609 JAM33106.1 GDIQ01157919 JAK93806.1 GDIP01124006 JAL79708.1 GDIQ01170919 JAK80806.1 GDIQ01131435 JAL20291.1 GDIP01158836 JAJ64566.1 GDIP01138351 JAL65363.1 GDIQ01167071 JAK84654.1 GDIQ01041668 JAN53069.1 GDIQ01106643 JAL45083.1 GDIQ01090646 JAN04091.1 GFDF01002705 JAV11379.1 GFDF01002706 JAV11378.1
KOB71324.1 ODYU01007549 SOQ50412.1 AGBW02008966 OWR52051.1 KQ460736 KPJ12601.1 KQ459463 KPJ00426.1 CCAG010000199 JRES01000321 KNC32294.1 CH902617 EDV42397.1 KK852832 KDR15313.1 GEZM01061243 JAV70608.1 JXJN01015477 CP012526 ALC47440.1 KA647220 AFP61849.1 APGK01039349 APGK01039350 BT127750 KB740970 AEE62712.1 ENN76643.1 KB632388 ERL94372.1 ATLV01019085 KE525262 KFB43588.1 GANO01003475 JAB56396.1 CH964232 EDW81204.1 OUUW01000007 SPP82730.1 CM000070 EAL27945.1 CH479179 EDW24444.1 GAPW01002363 JAC11235.1 GAMC01014978 JAB91577.1 GFDL01003242 JAV31803.1 GFDL01003240 JAV31805.1 GFDL01003300 JAV31745.1 GFDL01003306 JAV31739.1 CH933806 EDW15104.1 GAKP01004441 JAC54511.1 CH477648 EAT37827.1 CM000364 EDX14153.1 CM000160 EDW99013.1 UFQS01000083 UFQT01000083 SSW99069.1 SSX19451.1 CH940656 EDW58620.1 CVRI01000057 CRL02264.1 GGFM01004149 MBW24900.1 ADMH02002101 ETN59143.1 AE014297 AY121648 AAF56228.1 AAM51975.1 GDHF01001353 JAI50961.1 JXUM01073817 JXUM01073818 JXUM01073819 KQ562796 KXJ75113.1 CH480815 EDW43423.1 CH954182 EDV53923.1 NNAY01001807 OXU22866.1 AXCM01002705 AXCN02001502 GALX01002500 JAB65966.1 GBXI01017117 JAC97174.1 BT099602 ACU45755.1 KQ971311 KYB29477.1 GGFK01010561 MBW43882.1 CH916369 EDV93057.1 AAAB01008987 EAA01787.4 EGK97628.1 APCN01000826 AJWK01013903 NEVH01006990 PNF36186.1 GDIQ01033973 JAN60764.1 GDIQ01153078 JAK98647.1 GFDF01002532 JAV11552.1 CAEY01001378 GDIQ01153079 JAK98646.1 GDIP01058605 JAM45110.1 GDIQ01218256 JAK33469.1 GDIQ01155777 JAK95948.1 GDIP01070609 JAM33106.1 GDIQ01157919 JAK93806.1 GDIP01124006 JAL79708.1 GDIQ01170919 JAK80806.1 GDIQ01131435 JAL20291.1 GDIP01158836 JAJ64566.1 GDIP01138351 JAL65363.1 GDIQ01167071 JAK84654.1 GDIQ01041668 JAN53069.1 GDIQ01106643 JAL45083.1 GDIQ01090646 JAN04091.1 GFDF01002705 JAV11379.1 GFDF01002706 JAV11378.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053240
UP000053268
+ More
UP000092444 UP000037069 UP000007801 UP000027135 UP000092443 UP000092460 UP000092553 UP000095301 UP000019118 UP000030742 UP000030765 UP000007798 UP000078200 UP000268350 UP000001819 UP000095300 UP000008744 UP000075880 UP000009192 UP000008820 UP000000304 UP000002282 UP000091820 UP000008792 UP000183832 UP000192221 UP000002358 UP000000673 UP000075885 UP000000803 UP000069940 UP000249989 UP000001292 UP000008711 UP000215335 UP000075883 UP000075886 UP000075884 UP000007266 UP000001070 UP000007062 UP000076407 UP000075900 UP000075903 UP000075902 UP000069272 UP000076408 UP000075840 UP000075882 UP000075881 UP000092461 UP000235965 UP000015104
UP000092444 UP000037069 UP000007801 UP000027135 UP000092443 UP000092460 UP000092553 UP000095301 UP000019118 UP000030742 UP000030765 UP000007798 UP000078200 UP000268350 UP000001819 UP000095300 UP000008744 UP000075880 UP000009192 UP000008820 UP000000304 UP000002282 UP000091820 UP000008792 UP000183832 UP000192221 UP000002358 UP000000673 UP000075885 UP000000803 UP000069940 UP000249989 UP000001292 UP000008711 UP000215335 UP000075883 UP000075886 UP000075884 UP000007266 UP000001070 UP000007062 UP000076407 UP000075900 UP000075903 UP000075902 UP000069272 UP000076408 UP000075840 UP000075882 UP000075881 UP000092461 UP000235965 UP000015104
Interpro
SUPFAM
SSF52821
SSF52821
Gene 3D
ProteinModelPortal
H9IWY3
A0A2A4JCW4
A0A2W1BHW7
A0A0L7L7K7
A0A2H1WC49
A0A212FE93
+ More
A0A0N0PC89 A0A194QAI4 A0A1B0G858 A0A0L0CLH3 B3M2S6 A0A067R9S4 A0A1Y1LCI2 A0A1A9XI27 A0A1B0BJP7 A0A0M4EP02 A0A1I8NB19 T1PGF3 J3JWY2 U4UMN9 A0A084W044 U5ES05 B4NBP0 A0A1A9VR18 A0A3B0K428 Q298L0 A0A1I8PGN0 B4G433 A0A023EQ94 A0A182J586 W8AZA7 A0A1Q3FWK8 A0A1Q3FW75 A0A1Q3FW18 A0A1Q3FVW1 B4K5A4 A0A034WFZ6 Q16TJ5 B4QSF4 B4PL28 A0A336LR80 A0A1A9X1B4 B4MBZ9 A0A1J1IQ26 A0A1W4V4T4 A0A1S4FPW9 K7IMK1 A0A2M3Z8T6 W5J6W7 A0A182PRT9 Q9VCE0 A0A0K8WII1 A0A182GM50 B4HGH0 B3P7A3 A0A232EWZ2 A0A182M5N0 A0A182QSB2 V5I9R0 A0A0A1WFB6 A0A182NQQ8 C6TPB6 A0A139WNM4 A0A2M4AT34 B4JIH6 Q7PX64 A0A182X9E5 A0A182RSB8 A0A182VHL7 A0A182TKY9 A0A087ZIK7 A0A182FS14 A0A182YHY5 A0A1Y9IXW8 A0A182HKD1 A0A182L8L6 A0A182JUS1 A0A1B0CIY5 A0A2J7R5S6 A0A0P6GF40 A0A0P5NGP9 A0A1L8DZ44 T1K3B0 A0A0P5MRD9 A0A0P5ZC79 A0A0P5JXN8 A0A0P5MJB2 A0A0P5XLG0 A0A0P5MBV6 A0A0P5TN97 A0A0P5M1H6 A0A0N8BYV3 A0A0P5DE76 A0A0N8CEY7 A0A0P5LPF9 A0A0N8EC00 A0A0N8C7Q2 A0A0P6DLD2 A0A1L8DY97 A0A1L8DY52
A0A0N0PC89 A0A194QAI4 A0A1B0G858 A0A0L0CLH3 B3M2S6 A0A067R9S4 A0A1Y1LCI2 A0A1A9XI27 A0A1B0BJP7 A0A0M4EP02 A0A1I8NB19 T1PGF3 J3JWY2 U4UMN9 A0A084W044 U5ES05 B4NBP0 A0A1A9VR18 A0A3B0K428 Q298L0 A0A1I8PGN0 B4G433 A0A023EQ94 A0A182J586 W8AZA7 A0A1Q3FWK8 A0A1Q3FW75 A0A1Q3FW18 A0A1Q3FVW1 B4K5A4 A0A034WFZ6 Q16TJ5 B4QSF4 B4PL28 A0A336LR80 A0A1A9X1B4 B4MBZ9 A0A1J1IQ26 A0A1W4V4T4 A0A1S4FPW9 K7IMK1 A0A2M3Z8T6 W5J6W7 A0A182PRT9 Q9VCE0 A0A0K8WII1 A0A182GM50 B4HGH0 B3P7A3 A0A232EWZ2 A0A182M5N0 A0A182QSB2 V5I9R0 A0A0A1WFB6 A0A182NQQ8 C6TPB6 A0A139WNM4 A0A2M4AT34 B4JIH6 Q7PX64 A0A182X9E5 A0A182RSB8 A0A182VHL7 A0A182TKY9 A0A087ZIK7 A0A182FS14 A0A182YHY5 A0A1Y9IXW8 A0A182HKD1 A0A182L8L6 A0A182JUS1 A0A1B0CIY5 A0A2J7R5S6 A0A0P6GF40 A0A0P5NGP9 A0A1L8DZ44 T1K3B0 A0A0P5MRD9 A0A0P5ZC79 A0A0P5JXN8 A0A0P5MJB2 A0A0P5XLG0 A0A0P5MBV6 A0A0P5TN97 A0A0P5M1H6 A0A0N8BYV3 A0A0P5DE76 A0A0N8CEY7 A0A0P5LPF9 A0A0N8EC00 A0A0N8C7Q2 A0A0P6DLD2 A0A1L8DY97 A0A1L8DY52
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Mitochondrion inner membrane
Anchored to the mitochondrial inner membrane through its interaction with the integral membrane beta chain. With evidence from 1 publications.
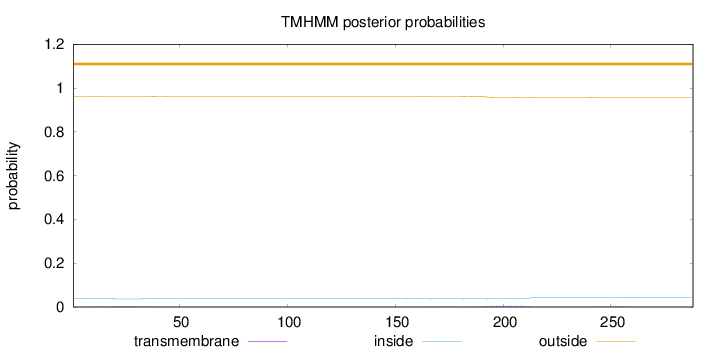
Length:
288
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.26677
Exp number, first 60 AAs:
0.07917
Total prob of N-in:
0.03807
outside
1 - 288
Population Genetic Test Statistics
Pi
149.950233
Theta
146.648618
Tajima's D
-0.646739
CLR
154.788129
CSRT
0.205339733013349
Interpretation
Uncertain
