Pre Gene Modal
BGIBMGA001670
Annotation
PREDICTED:_cytoplasmic_FMR1-interacting_protein_[Papilio_xuthus]
Full name
Cytoplasmic FMR1-interacting protein
Alternative Name
Specifically Rac1-associated protein 1
Location in the cell
Nuclear Reliability : 1.914
Sequence
CDS
ATGTCTGTAAGTGAAAAAGTGTCGTTATCGGATGCTCTGAGCAACGTAGATGTGTTGGATGAGCTGACTTTGCCGGATGAACAGCCATGTATCGAAGCAGCACCTTGCTCGATCCTGTATCAAGCTAATTTCGATACTAACTTTGAAGACCGCAATGGGTTCATCACCGGTATCGCAAAGTACATAGAGGAAGCGACCGTCCACGCCAACTTGAACGAGCTCCTGGAAGAAGGGAACGCCCACGCTGTCATGCTGTACACGTGGCGTTGCTGCAGCCGTGCCATACCTCAGCCCAGGTCCAATGAACAACCAGACCGGGTTCACATATACGAGAGGACCGTTCAAGTGCTGGCTCCTGAGGTTGATAAGCTCTTGCAGTTCATGTATTTCCAGCGCAAGGCCATAGAAAGGTTCTGTGCTGAAGTGCGTCGGCTGTGCCACGCGGAGAAGAGGAGGGACTTCGTCTCCGAAGCCTACCTCCTGACCCTCGGCAAGTTCGTGAACATGTTCGCGGTCCTCGACGAGCTGAAGAACATGAAGAGTTCGGTCAAGAATGATTACTCGACTTACAGAAGGGCGGCGCAATTCCTGAAGGTGATGTCCGACAGTCAGAGCCTGCAGGAGTCTCAGAACCTGTCCATGTTCCTCGCGACCCAGAACAAGATCAGAGACACGGTCAAGGATGCCCTAGAGAAAATTAACGGTTACGAAGACTTGTTGGCTGATGTGGTCAATATATGCGTGCACATGTTCGAGACAAGAATGTATTTGACTCCATGCGAGAAACACATGCTCGTGAAGGTTATGGGCTTCGGACTCTTCCTCATGGACTCAGAGGTGTGCAACATCAATCGTTTGGATCAGAAGAAGAAGATACGTCTGGACCGCATCGACAGGATCTTCAAGAACCTCGAAGTGGTTCCACTGTTTGGCGACATGCAAATAGCTCCTTTCAATTACATCAAGAGATCGAAACACTATGATCCGAATAAGTGGCCCCTGTCCTCGAGCCCCAACCCGCCCTCGCCGCAGGCCGACCTGATGGTCCACCTGCCCCAGATCCGGGAGGAGCACCAGAACTACATATCCGAGCTGGCCCGGTACAGCAACGAGGTGACGACCACTTTCAAAGAGGCGGGTTCGGACTCCGAGAACAAGGCGGTGACGGAGCTCTGTCTGCGCGGCCTGCAGCTGCTGTCCTCCTGGTGCTCGGTGCTCACGGAGCTGTGCTCGTGGAAGCTGCTCCACCCGACGGACCACACCATCAACGCGCGGTGTCCCCCCGAAGCCGAGGAGTACGAGAGGGCGACGCGTTACAACTACACGTCCGAGGAGAAGTTCGCCATGATCGAGGTGATCGCGATGATAAAGGGCCTCCAGGTCCTGATGGCGCGGATGGAGACCGTGTTCGCGGACGCGGCCAGGAGGTCCATATACGCGGAGCTGCAGGACTTCGTGCAGCTCGTCCTGCGGGAGCCCCTGAGGAAGGCCATCAAGAACAAGAAGGACCTCATCCGGAGCATAATAGTGTCGGTTCGGGAGACGTGTGGTGACTGGGCGCGCGGCTGCGAGCCTCAGCAGGACCCGGCGCTCAAGGGGAAGAAGGACGGCGAAGCCAGCTTCACCATCAAGGTGCCCAGAAGGAATGTCGGTCCCTCATCGACCCAGCTCTACATGGTGAGAACGCAGCTTGAGGCCCTCATATCGGACAAGTCCGGAGGCCGGCGGACTCTTCGGAAAGACCTGGACGCAGCCACCCTGCATCAGATCGAAAGCTTCCACAGAACCAGCTTTTATTGGTCCTACCTGCTGAACTTACCTGATTCCCTGCAGAAGTGCTGCGATCTGTCTCAACTCTGGTATAGAGAGTTTTATCTCGAAATGACAATGGGAAAGAAAGTTAATAAATGCATGGTGCGTCATCAGCACAACGAGGAGTGCAACGACCTGGTCACGATGGAGAAGCGGATACAGTTCCCCATCGAGATGTCCATGCCATGGATATTGACCGAGCACATATTAAGGACCAAAGAACCGGCGATGATGGAATACGTCCTGTACCCGCTGGACCTGTACAACGACTCGGCCCAGTACGCGCTGACCGTCTTCAAGAAGCAGTTCCTCTACGACGAGGTGGAGGCGGAAGTCAACCTCTGCTTCGATCAGTTCGTGTACAAGCTCTCCGAGCAGGTCTACGCGTACTACAAGCAGCTGGCCGCGAGCATGCTGCTGGATAAGCGCTACCGTGCCGAGTGCTTGTCGCGAGGCGCGGGCGTCGGTGTCGGTCCAGGCCGGTACGCCTCCTTACTCAAGCAGAGGCACGTGGCTCTACTGGGACGGCACGTCGACCTCTGCGCGCTGGTGGCGCAGCGTATCAACTCGGACATGCACCGCGCTCTGGACGCGGCCATCGCGAAGTTCGAGGCCGGGGACATCACCGGCGTGGTGGAGCTGGAGGGGCTGATCGCGGTGAACCGTCTCTGCCACAAGCTGCTCAGCCGCTACCTGACCTTGGACGACTTCGAGGCGGTGCTGCGGGAGTCGGACCACGGCGTGCTGGCCCCGTACGGCAGGGTCACGCTGCACGTCTTCTGGGAGCTCAACTACGACTTCCTGCCGAACTACTGCTACAACGCTGCGACAGCGCGGTTCGTGAAGTGTCGCGGGATCCAGTTCGCGGCCAACGTGCAGCGGGAGCGCCCGCAACAGTACGGGCACGCGCTGCTGTGGGGCTCCAAGCAGCTCAGCCTCGTGTACTCCGCGCAGTACACGCAGTACAACGGGTTCGTGGGGGCGGCGCACCTGCACGCGCTGGTCCGGCTGCTGGGCTACCAGGGCGTGGCCGTGGTCGTGGGCGAGCTGCTGGGTGTGGCCCGTGGACTGCTGCACGGCTCGCTGGCGCAGTTCACGCGGGCCCTGCTCGCCGCGCTGCCGCGACACTGCCGCCTGCCCAGATACGACTACGGCTCCAATGGCGTGCTGGGCTACTACCACGCGCAGCTGACGGACATCGTGCAGTACCCCGACGCGAAGACGGAACTCTTCCACGCCTTCAGGGAGCTCGGGAACATCATCCTGTTCTGCATGCTGATCGAGCAGGCTTTAAGCCAAGAGGAGGTGACGGATCTGCTCCACGCGGCGCCCTTCCAGAACATTCTGCCGCGTCCCTACAGCACAGTTGCAGACGGTGATAAGCCCGAATCAAAACAAAAACGTCTGGAGGCCAAGTACGCGTCCCTACAGATCGTCCATAATGTCGACAAGTACGGCACCGCCAAGCAAGGTCAGCTGTCCCGCGAGGGCGACCTGCTGACCCGCGAGCGGCTGTGCTGCGGCCTGTCGCTGTTCGGCGTGGTGCTGCGGCGGCTGCGCACCTGCCTCGCCGCGCCCGCCTGGCCCGCGCCCCCCGCCGCGCACCACGCGCACACCGACGACACCAACGAGTTCCACAGATTGTGGTCAGCCCTGCAGTTCCTGTACTGCATTCCGGTCGGCGACACCCAGTTTACCGTCGAGGAACTGTTCGGCGAGGGTCTCCATTGGGCCGGCTGCACCATCATCGCTCTCCTGGGGCAGCAGAGGAGGTTCGAGGCGCTGGACTTCTGCTACCACATCCTGAGGGTGCAGAGGGTCGACGGAAAGGATGAACTGGTCAAAGGCATTCCCCTCAAGCGGATGGTGGACCGCATCAGGCGGTTCCAAGTGCTGAACTCGCAGATCTTCGGCGTGCTGTCCAGGCAGCTGTCCGCGGACGACGCGGCCGGCGTCGAGCACGTGCGCTGCTTCCCGCCCCCGCAGGCGCCGCTTACGCTCAACTGA
Protein
MSVSEKVSLSDALSNVDVLDELTLPDEQPCIEAAPCSILYQANFDTNFEDRNGFITGIAKYIEEATVHANLNELLEEGNAHAVMLYTWRCCSRAIPQPRSNEQPDRVHIYERTVQVLAPEVDKLLQFMYFQRKAIERFCAEVRRLCHAEKRRDFVSEAYLLTLGKFVNMFAVLDELKNMKSSVKNDYSTYRRAAQFLKVMSDSQSLQESQNLSMFLATQNKIRDTVKDALEKINGYEDLLADVVNICVHMFETRMYLTPCEKHMLVKVMGFGLFLMDSEVCNINRLDQKKKIRLDRIDRIFKNLEVVPLFGDMQIAPFNYIKRSKHYDPNKWPLSSSPNPPSPQADLMVHLPQIREEHQNYISELARYSNEVTTTFKEAGSDSENKAVTELCLRGLQLLSSWCSVLTELCSWKLLHPTDHTINARCPPEAEEYERATRYNYTSEEKFAMIEVIAMIKGLQVLMARMETVFADAARRSIYAELQDFVQLVLREPLRKAIKNKKDLIRSIIVSVRETCGDWARGCEPQQDPALKGKKDGEASFTIKVPRRNVGPSSTQLYMVRTQLEALISDKSGGRRTLRKDLDAATLHQIESFHRTSFYWSYLLNLPDSLQKCCDLSQLWYREFYLEMTMGKKVNKCMVRHQHNEECNDLVTMEKRIQFPIEMSMPWILTEHILRTKEPAMMEYVLYPLDLYNDSAQYALTVFKKQFLYDEVEAEVNLCFDQFVYKLSEQVYAYYKQLAASMLLDKRYRAECLSRGAGVGVGPGRYASLLKQRHVALLGRHVDLCALVAQRINSDMHRALDAAIAKFEAGDITGVVELEGLIAVNRLCHKLLSRYLTLDDFEAVLRESDHGVLAPYGRVTLHVFWELNYDFLPNYCYNAATARFVKCRGIQFAANVQRERPQQYGHALLWGSKQLSLVYSAQYTQYNGFVGAAHLHALVRLLGYQGVAVVVGELLGVARGLLHGSLAQFTRALLAALPRHCRLPRYDYGSNGVLGYYHAQLTDIVQYPDAKTELFHAFRELGNIILFCMLIEQALSQEEVTDLLHAAPFQNILPRPYSTVADGDKPESKQKRLEAKYASLQIVHNVDKYGTAKQGQLSREGDLLTRERLCCGLSLFGVVLRRLRTCLAAPAWPAPPAAHHAHTDDTNEFHRLWSALQFLYCIPVGDTQFTVEELFGEGLHWAGCTIIALLGQQRRFEALDFCYHILRVQRVDGKDELVKGIPLKRMVDRIRRFQVLNSQIFGVLSRQLSADDAAGVEHVRCFPPPQAPLTLN
Summary
Description
Plays a role in guidance and morphology of central and peripheral axons and in synaptic morphology. Also required for formation of cell membrane protrusions and for bristle development (By similarity).
Plays a role in guidance and morphology of central and peripheral axons and in synaptic morphology. Also required for formation of cell membrane protrusions and for bristle development.
Plays a role in guidance and morphology of central and peripheral axons and in synaptic morphology. Also required for formation of cell membrane protrusions and for bristle development.
Subunit
Interacts with Fmr1 and Rac1. Component of the WAVE complex composed of Hem/Kette, Scar/Wave and Sra-1/Cyfip where it binds through its C-terminus directly to Hem (By similarity).
Interacts with Fmr1 and Rac1. Component of the WAVE complex composed of Hem/Kette, Scar/Wave and Sra-1/Cyfip where it binds through its C-terminus directly to Hem.
Interacts with Fmr1 and Rac1. Component of the WAVE complex composed of Hem/Kette, Scar/Wave and Sra-1/Cyfip where it binds through its C-terminus directly to Hem.
Similarity
Belongs to the CYFIP family.
Keywords
Cell shape
Complete proteome
Cytoplasm
Developmental protein
Differentiation
Neurogenesis
Reference proteome
Feature
chain Cytoplasmic FMR1-interacting protein
Uniprot
A0A212FEB2
H9IWN8
A0A1Y1LBS7
A0A1W4X9U0
A0A1W4X0D3
A0A0A9YMY3
+ More
A0A154PF73 A0A067QJN8 A0A087ZRN0 D6W6N5 A0A023F4Y3 A0A0M9A438 A0A2H8TTY5 J9JL00 A0A2S2PGC3 A0A069DZS4 A0A195BYR5 A0A195FLC5 A0A0C9RNX2 A0A026VST8 A0A195ENV3 F4WRU2 A0A195BVS2 A0A158NEL8 A0A3L8DDD8 E2AEJ6 A0A151XIS4 E9IJB2 T1I0M5 A0A0A9YLG4 E0VKF3 A0A182IU35 A0A1I8P265 E2BT38 A0A1L8DJK9 A0A0L0CRE7 A0A182FK33 A0A182W4Z5 W5JMR6 T1PGJ9 A0A182K353 A0A182P7I9 A0A1A9VLJ8 A0A1A9ZN40 A0A182V303 Q7QEX4 U5EXQ3 U4UVP4 A0A182MF32 A0A182R7H8 A0A182N1K0 A0A182Q9W1 B4N7Y7 A0A1A9X2J4 A0A182GY71 B4G548 B4HDX6 B3P0V0 Q299G2 Q9VF87 A0A182YQ59 A0A3B0KPH8 U3PXB4 A0A1Q3FXC0 W8AKB2 Q16ZZ3 A0A1W4VEF2 B4MBK0 B4KCY0 A0A0K8VKF5 B4PS48 A0A034VU41 A0A0M4EPP6 N6U0N1 A0A310SRG8 B3MSU9 A0A0A1X1G2 A0A084WPX5 A0A3S2MA46 B4JS07 A0A182HIB1 A0A3B0K0E4 A0A336KRU0 K7IU01 A0A1B6IXR2 A0A1J1IGB3 A0A0P5JWY7 E9G9J2 A0A0P4Z605 A0A0P6A2V9 W8B750 A0A0P5YDJ2 A0A0P5MU60 A0A1B6J2I2 A0A0P5JIL9 A0A226E545 A0A2J7R5M9 B4QYJ2 A0A0P4YFE5 A0A182X808
A0A154PF73 A0A067QJN8 A0A087ZRN0 D6W6N5 A0A023F4Y3 A0A0M9A438 A0A2H8TTY5 J9JL00 A0A2S2PGC3 A0A069DZS4 A0A195BYR5 A0A195FLC5 A0A0C9RNX2 A0A026VST8 A0A195ENV3 F4WRU2 A0A195BVS2 A0A158NEL8 A0A3L8DDD8 E2AEJ6 A0A151XIS4 E9IJB2 T1I0M5 A0A0A9YLG4 E0VKF3 A0A182IU35 A0A1I8P265 E2BT38 A0A1L8DJK9 A0A0L0CRE7 A0A182FK33 A0A182W4Z5 W5JMR6 T1PGJ9 A0A182K353 A0A182P7I9 A0A1A9VLJ8 A0A1A9ZN40 A0A182V303 Q7QEX4 U5EXQ3 U4UVP4 A0A182MF32 A0A182R7H8 A0A182N1K0 A0A182Q9W1 B4N7Y7 A0A1A9X2J4 A0A182GY71 B4G548 B4HDX6 B3P0V0 Q299G2 Q9VF87 A0A182YQ59 A0A3B0KPH8 U3PXB4 A0A1Q3FXC0 W8AKB2 Q16ZZ3 A0A1W4VEF2 B4MBK0 B4KCY0 A0A0K8VKF5 B4PS48 A0A034VU41 A0A0M4EPP6 N6U0N1 A0A310SRG8 B3MSU9 A0A0A1X1G2 A0A084WPX5 A0A3S2MA46 B4JS07 A0A182HIB1 A0A3B0K0E4 A0A336KRU0 K7IU01 A0A1B6IXR2 A0A1J1IGB3 A0A0P5JWY7 E9G9J2 A0A0P4Z605 A0A0P6A2V9 W8B750 A0A0P5YDJ2 A0A0P5MU60 A0A1B6J2I2 A0A0P5JIL9 A0A226E545 A0A2J7R5M9 B4QYJ2 A0A0P4YFE5 A0A182X808
Pubmed
22118469
19121390
28004739
25401762
26823975
24845553
+ More
18362917 19820115 25474469 26334808 24508170 21719571 21347285 30249741 20798317 21282665 20566863 26108605 20920257 23761445 25315136 12364791 14747013 17210077 23537049 17994087 26483478 15632085 11438699 10731132 12537572 12537569 14588242 12818175 15385157 15269173 25244985 24495485 17510324 17550304 25348373 25830018 24438588 20075255 21292972
18362917 19820115 25474469 26334808 24508170 21719571 21347285 30249741 20798317 21282665 20566863 26108605 20920257 23761445 25315136 12364791 14747013 17210077 23537049 17994087 26483478 15632085 11438699 10731132 12537572 12537569 14588242 12818175 15385157 15269173 25244985 24495485 17510324 17550304 25348373 25830018 24438588 20075255 21292972
EMBL
AGBW02008966
OWR52053.1
BABH01021128
BABH01021129
BABH01021130
BABH01021131
+ More
BABH01021132 BABH01021133 GEZM01060282 JAV71122.1 GBHO01011146 GDHC01017156 JAG32458.1 JAQ01473.1 KQ434886 KZC10094.1 KK853265 KDR09197.1 KQ971307 EFA10974.1 GBBI01002156 JAC16556.1 KQ435740 KOX76825.1 GFXV01005918 MBW17723.1 ABLF02026358 GGMR01015860 MBY28479.1 GBGD01000135 JAC88754.1 KQ978501 KYM93455.1 KQ981490 KYN41213.1 GBYB01010140 JAG79907.1 KK108171 EZA46838.1 KQ978625 KYN29579.1 GL888292 EGI63099.1 KQ976401 KYM92370.1 ADTU01013497 QOIP01000009 RLU18467.1 GL438862 EFN68153.1 KQ982080 KYQ60306.1 GL763764 EFZ19330.1 ACPB03016920 ACPB03016921 ACPB03016922 ACPB03016923 GBHO01011148 JAG32456.1 DS235243 EEB13869.1 GL450325 EFN81168.1 GFDF01007529 JAV06555.1 JRES01000007 KNC34935.1 ADMH02000604 ETN65677.1 KA647906 AFP62535.1 AAAB01008846 EAA06404.3 GANO01000835 JAB59036.1 KB632384 ERL94290.1 AXCM01000253 AXCN02002442 CH964232 EDW81238.1 JXUM01096983 JXUM01096984 JXUM01096985 JXUM01096986 KQ564307 KXJ72452.1 CH479179 EDW24714.1 CH480815 EDW42068.1 CH954181 EDV48998.1 CM000070 EAL27741.1 AY017343 AY029211 AE014297 BT011115 AY122203 AAF55173.1 AAG61254.1 AAK31584.1 AAM52715.1 AAR82782.1 OUUW01000013 SPP87776.1 BT150379 AGW52189.1 GFDL01002801 JAV32244.1 GAMC01017505 JAB89050.1 CH477480 EAT40231.1 CH940656 EDW58471.1 CH933806 EDW17002.1 GDHF01027818 GDHF01013264 JAI24496.1 JAI39050.1 CM000160 EDW97468.2 GAKP01013894 JAC45058.1 CP012526 ALC47790.1 APGK01047141 KB741077 ENN74156.1 KQ759874 OAD62357.1 CH902623 EDV30339.1 GBXI01009360 JAD04932.1 ATLV01025136 KE525369 KFB52269.1 RSAL01000002 RVE54909.1 CH916373 EDV94547.1 APCN01003925 SPP87777.1 UFQS01000761 UFQS01000777 UFQT01000761 UFQT01000777 SSX06699.1 SSX06812.1 SSX27045.1 GECU01015979 JAS91727.1 CVRI01000050 CRK99285.1 GDIQ01197841 GDIQ01027653 JAK53884.1 GL732536 EFX83871.1 GDIP01229363 JAI94038.1 GDIP01036484 JAM67231.1 GAMC01017504 JAB89051.1 GDIP01059232 JAM44483.1 GDIQ01159253 JAK92472.1 GECU01014312 JAS93394.1 GDIQ01197842 JAK53883.1 LNIX01000006 OXA52418.1 NEVH01006991 PNF36132.1 CM000364 EDX12837.1 GDIP01229364 JAI94037.1
BABH01021132 BABH01021133 GEZM01060282 JAV71122.1 GBHO01011146 GDHC01017156 JAG32458.1 JAQ01473.1 KQ434886 KZC10094.1 KK853265 KDR09197.1 KQ971307 EFA10974.1 GBBI01002156 JAC16556.1 KQ435740 KOX76825.1 GFXV01005918 MBW17723.1 ABLF02026358 GGMR01015860 MBY28479.1 GBGD01000135 JAC88754.1 KQ978501 KYM93455.1 KQ981490 KYN41213.1 GBYB01010140 JAG79907.1 KK108171 EZA46838.1 KQ978625 KYN29579.1 GL888292 EGI63099.1 KQ976401 KYM92370.1 ADTU01013497 QOIP01000009 RLU18467.1 GL438862 EFN68153.1 KQ982080 KYQ60306.1 GL763764 EFZ19330.1 ACPB03016920 ACPB03016921 ACPB03016922 ACPB03016923 GBHO01011148 JAG32456.1 DS235243 EEB13869.1 GL450325 EFN81168.1 GFDF01007529 JAV06555.1 JRES01000007 KNC34935.1 ADMH02000604 ETN65677.1 KA647906 AFP62535.1 AAAB01008846 EAA06404.3 GANO01000835 JAB59036.1 KB632384 ERL94290.1 AXCM01000253 AXCN02002442 CH964232 EDW81238.1 JXUM01096983 JXUM01096984 JXUM01096985 JXUM01096986 KQ564307 KXJ72452.1 CH479179 EDW24714.1 CH480815 EDW42068.1 CH954181 EDV48998.1 CM000070 EAL27741.1 AY017343 AY029211 AE014297 BT011115 AY122203 AAF55173.1 AAG61254.1 AAK31584.1 AAM52715.1 AAR82782.1 OUUW01000013 SPP87776.1 BT150379 AGW52189.1 GFDL01002801 JAV32244.1 GAMC01017505 JAB89050.1 CH477480 EAT40231.1 CH940656 EDW58471.1 CH933806 EDW17002.1 GDHF01027818 GDHF01013264 JAI24496.1 JAI39050.1 CM000160 EDW97468.2 GAKP01013894 JAC45058.1 CP012526 ALC47790.1 APGK01047141 KB741077 ENN74156.1 KQ759874 OAD62357.1 CH902623 EDV30339.1 GBXI01009360 JAD04932.1 ATLV01025136 KE525369 KFB52269.1 RSAL01000002 RVE54909.1 CH916373 EDV94547.1 APCN01003925 SPP87777.1 UFQS01000761 UFQS01000777 UFQT01000761 UFQT01000777 SSX06699.1 SSX06812.1 SSX27045.1 GECU01015979 JAS91727.1 CVRI01000050 CRK99285.1 GDIQ01197841 GDIQ01027653 JAK53884.1 GL732536 EFX83871.1 GDIP01229363 JAI94038.1 GDIP01036484 JAM67231.1 GAMC01017504 JAB89051.1 GDIP01059232 JAM44483.1 GDIQ01159253 JAK92472.1 GECU01014312 JAS93394.1 GDIQ01197842 JAK53883.1 LNIX01000006 OXA52418.1 NEVH01006991 PNF36132.1 CM000364 EDX12837.1 GDIP01229364 JAI94037.1
Proteomes
UP000007151
UP000005204
UP000192223
UP000076502
UP000027135
UP000005203
+ More
UP000007266 UP000053105 UP000007819 UP000078542 UP000078541 UP000053097 UP000078492 UP000007755 UP000078540 UP000005205 UP000279307 UP000000311 UP000075809 UP000015103 UP000009046 UP000075880 UP000095300 UP000008237 UP000037069 UP000069272 UP000075920 UP000000673 UP000095301 UP000075881 UP000075885 UP000078200 UP000092445 UP000075903 UP000007062 UP000030742 UP000075883 UP000075900 UP000075884 UP000075886 UP000007798 UP000091820 UP000069940 UP000249989 UP000008744 UP000001292 UP000008711 UP000001819 UP000000803 UP000076408 UP000268350 UP000008820 UP000192221 UP000008792 UP000009192 UP000002282 UP000092553 UP000019118 UP000007801 UP000030765 UP000283053 UP000001070 UP000075840 UP000002358 UP000183832 UP000000305 UP000198287 UP000235965 UP000000304 UP000076407
UP000007266 UP000053105 UP000007819 UP000078542 UP000078541 UP000053097 UP000078492 UP000007755 UP000078540 UP000005205 UP000279307 UP000000311 UP000075809 UP000015103 UP000009046 UP000075880 UP000095300 UP000008237 UP000037069 UP000069272 UP000075920 UP000000673 UP000095301 UP000075881 UP000075885 UP000078200 UP000092445 UP000075903 UP000007062 UP000030742 UP000075883 UP000075900 UP000075884 UP000075886 UP000007798 UP000091820 UP000069940 UP000249989 UP000008744 UP000001292 UP000008711 UP000001819 UP000000803 UP000076408 UP000268350 UP000008820 UP000192221 UP000008792 UP000009192 UP000002282 UP000092553 UP000019118 UP000007801 UP000030765 UP000283053 UP000001070 UP000075840 UP000002358 UP000183832 UP000000305 UP000198287 UP000235965 UP000000304 UP000076407
Interpro
ProteinModelPortal
A0A212FEB2
H9IWN8
A0A1Y1LBS7
A0A1W4X9U0
A0A1W4X0D3
A0A0A9YMY3
+ More
A0A154PF73 A0A067QJN8 A0A087ZRN0 D6W6N5 A0A023F4Y3 A0A0M9A438 A0A2H8TTY5 J9JL00 A0A2S2PGC3 A0A069DZS4 A0A195BYR5 A0A195FLC5 A0A0C9RNX2 A0A026VST8 A0A195ENV3 F4WRU2 A0A195BVS2 A0A158NEL8 A0A3L8DDD8 E2AEJ6 A0A151XIS4 E9IJB2 T1I0M5 A0A0A9YLG4 E0VKF3 A0A182IU35 A0A1I8P265 E2BT38 A0A1L8DJK9 A0A0L0CRE7 A0A182FK33 A0A182W4Z5 W5JMR6 T1PGJ9 A0A182K353 A0A182P7I9 A0A1A9VLJ8 A0A1A9ZN40 A0A182V303 Q7QEX4 U5EXQ3 U4UVP4 A0A182MF32 A0A182R7H8 A0A182N1K0 A0A182Q9W1 B4N7Y7 A0A1A9X2J4 A0A182GY71 B4G548 B4HDX6 B3P0V0 Q299G2 Q9VF87 A0A182YQ59 A0A3B0KPH8 U3PXB4 A0A1Q3FXC0 W8AKB2 Q16ZZ3 A0A1W4VEF2 B4MBK0 B4KCY0 A0A0K8VKF5 B4PS48 A0A034VU41 A0A0M4EPP6 N6U0N1 A0A310SRG8 B3MSU9 A0A0A1X1G2 A0A084WPX5 A0A3S2MA46 B4JS07 A0A182HIB1 A0A3B0K0E4 A0A336KRU0 K7IU01 A0A1B6IXR2 A0A1J1IGB3 A0A0P5JWY7 E9G9J2 A0A0P4Z605 A0A0P6A2V9 W8B750 A0A0P5YDJ2 A0A0P5MU60 A0A1B6J2I2 A0A0P5JIL9 A0A226E545 A0A2J7R5M9 B4QYJ2 A0A0P4YFE5 A0A182X808
A0A154PF73 A0A067QJN8 A0A087ZRN0 D6W6N5 A0A023F4Y3 A0A0M9A438 A0A2H8TTY5 J9JL00 A0A2S2PGC3 A0A069DZS4 A0A195BYR5 A0A195FLC5 A0A0C9RNX2 A0A026VST8 A0A195ENV3 F4WRU2 A0A195BVS2 A0A158NEL8 A0A3L8DDD8 E2AEJ6 A0A151XIS4 E9IJB2 T1I0M5 A0A0A9YLG4 E0VKF3 A0A182IU35 A0A1I8P265 E2BT38 A0A1L8DJK9 A0A0L0CRE7 A0A182FK33 A0A182W4Z5 W5JMR6 T1PGJ9 A0A182K353 A0A182P7I9 A0A1A9VLJ8 A0A1A9ZN40 A0A182V303 Q7QEX4 U5EXQ3 U4UVP4 A0A182MF32 A0A182R7H8 A0A182N1K0 A0A182Q9W1 B4N7Y7 A0A1A9X2J4 A0A182GY71 B4G548 B4HDX6 B3P0V0 Q299G2 Q9VF87 A0A182YQ59 A0A3B0KPH8 U3PXB4 A0A1Q3FXC0 W8AKB2 Q16ZZ3 A0A1W4VEF2 B4MBK0 B4KCY0 A0A0K8VKF5 B4PS48 A0A034VU41 A0A0M4EPP6 N6U0N1 A0A310SRG8 B3MSU9 A0A0A1X1G2 A0A084WPX5 A0A3S2MA46 B4JS07 A0A182HIB1 A0A3B0K0E4 A0A336KRU0 K7IU01 A0A1B6IXR2 A0A1J1IGB3 A0A0P5JWY7 E9G9J2 A0A0P4Z605 A0A0P6A2V9 W8B750 A0A0P5YDJ2 A0A0P5MU60 A0A1B6J2I2 A0A0P5JIL9 A0A226E545 A0A2J7R5M9 B4QYJ2 A0A0P4YFE5 A0A182X808
PDB
4N78
E-value=0,
Score=4076
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Accumulates in central axons and motor neuron terminals. With evidence from 1 publications.
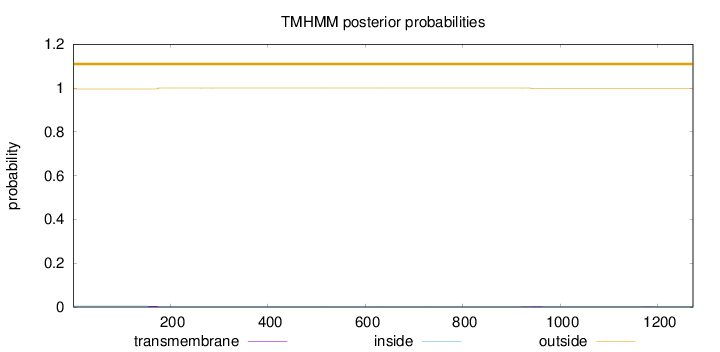
Length:
1272
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.150020000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00453
outside
1 - 1272
Population Genetic Test Statistics
Pi
199.741292
Theta
160.597015
Tajima's D
0.876079
CLR
0.419724
CSRT
0.625668716564172
Interpretation
Uncertain
