Gene
KWMTBOMO06166
Pre Gene Modal
BGIBMGA001763
Annotation
PREDICTED:_uncharacterized_protein_LOC101735722_[Bombyx_mori]
Location in the cell
ER Reliability : 2.016
Sequence
CDS
ATGATCGCCCTGAAGAAACTATATGGCGACGAGCTACTTCGCCTGGCTCTGGTTTTGAGTTTGCTCAGGATCGACCCAGACGACTACCTCGAACGGGAGACCCAGTTGCTGGCCGGTCTGCACATCGCGAACGGTTCCGACCTGTCCCCGGAACAGGAGGCCCGGACGCTGATACGGCCGAGGTTCGTCCACCCCAAGTCCCTGGACAACATCCTCGTGAACTACGAGAGGCAGCTGTTTGAAGAGCTCAACTCCTTGGGCCGGTACGGTCCAAGCGCACATAATGCCCGGATGTGGCAGAACCGTGGCCCGATTCTTCATACATATTTATTGAACGATGTCGCAACTGGCGCAGTCGAACCCGTGTCAGCGGCCCAGTCGGTCGCAACCGACCAGGACGCCGTCTCGGCGGACGACAACGTCGCCCAGGAGGTCAAAGTGACCGACGAGGACGCCCAAGACCAAAGTCGCGAGTCCACGGTGACGGTGTCGGACAACTTCCTCCACAATCGCACCGAATCGGATATATTTCCGGAGATCGCTTCCAGAACCTTCGACGTGACCGAGTTCCTGGTGCAGCAGGACAATATGCAAATTAAAAAGGAACAGGACGAGGCTTTCGACCTCAAGGTCAACGAATCGAACACCGATGACTTTTACAGTGACAACGCTATCGACTTTGCGACTTATTTTGCTGCAAGATCCGAGAAGATCGAGGTGGAGGAGCCTAATTTATATCACCAGAATATGGCCGACCTTCAAGATTATGGGGACGTCTTCACCGACGATCCGGACCTCAAACACGACCTGGAACTAGACATAAAACAACAGCAGGAGAACTTGGAACAGTATCTCCTTGAGAACACTCCTTTGGAAGCGGAGGTCTATGAATTGGCTCCAATGTTTGAAGAGGAACTGGTGTTGGACGTTAAGAGGGAAAGAGCTAGCACTAGTTTCACGTCGGCCAGTTCGAGTGGCGTGAGCGAGATGGACATATCCGACGTCAAAACCGAGCCGTTGGACTATAATGACGTCAAGGTCGAGCCTGATGAGGGGAACCATACTGGAGATGAACTGACGCAAGAGGACATGGATCTCATCGAGGTGCTCTGGAAGCAGGACGTGGACATGGGATTCTCGCTCGAAGATCCAATACAGCCGGAAGGTCCGCAAGCCACGAAGGTATTGGGGGAGGAAATCGACAAGGAACTGCTGAAGGCCAAAGATAGGATCCAGAAGAACTATGAAGATGAAGAGCTGAAGAAGGTTGAGATAGAGAAGGTCAAGGCTTCGTTGTTGGATCCCGAAGTCAAAGAGGATGATCCCTGGGCTGGCCTATCCTACACCGTCGACACGGAAACCGGCGAATACGTGATCCAAGGAGAACTGCCCAGGGAGCTGGTGAGCGGCGAGGAGTCGCGCTTCGATTTGCTGGAGGAGGCGCTTCAGCTGGTGGAACTCGGAGACGGCGCTGAAGCCAAAGATGAACCCACTGAGGCGGTAGAAGGGAGTAGCACCGATCGCAGGAGCACCATGTTGCACCCTGCCATGCGGCATGTCCCGCACCACCCCCTCGCCCATTACCACAACCAGAACAGCGAGATTACGTCGCAGCCAATATTTATTTGA
Protein
MIALKKLYGDELLRLALVLSLLRIDPDDYLERETQLLAGLHIANGSDLSPEQEARTLIRPRFVHPKSLDNILVNYERQLFEELNSLGRYGPSAHNARMWQNRGPILHTYLLNDVATGAVEPVSAAQSVATDQDAVSADDNVAQEVKVTDEDAQDQSRESTVTVSDNFLHNRTESDIFPEIASRTFDVTEFLVQQDNMQIKKEQDEAFDLKVNESNTDDFYSDNAIDFATYFAARSEKIEVEEPNLYHQNMADLQDYGDVFTDDPDLKHDLELDIKQQQENLEQYLLENTPLEAEVYELAPMFEEELVLDVKRERASTSFTSASSSGVSEMDISDVKTEPLDYNDVKVEPDEGNHTGDELTQEDMDLIEVLWKQDVDMGFSLEDPIQPEGPQATKVLGEEIDKELLKAKDRIQKNYEDEELKKVEIEKVKASLLDPEVKEDDPWAGLSYTVDTETGEYVIQGELPRELVSGEESRFDLLEEALQLVELGDGAEAKDEPTEAVEGSSTDRRSTMLHPAMRHVPHHPLAHYHNQNSEITSQPIFI
Summary
Similarity
Belongs to the bZIP family.
Uniprot
EMBL
Proteomes
Pfam
PF03131 bZIP_Maf
SUPFAM
SSF47454
SSF47454
ProteinModelPortal
Ontologies
KEGG
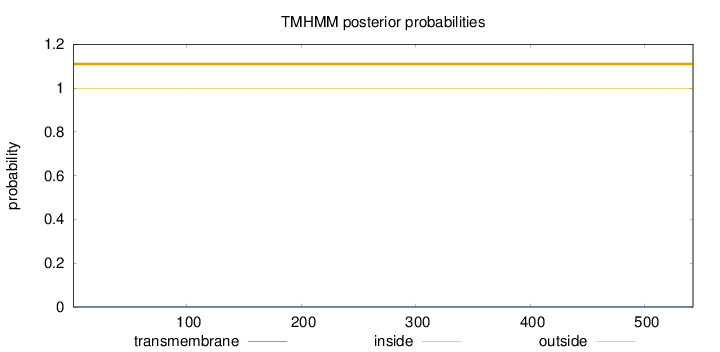
Topology
Subcellular location
Nucleus
Length:
542
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00202
Exp number, first 60 AAs:
0.00157
Total prob of N-in:
0.00117
outside
1 - 542
Population Genetic Test Statistics
Pi
271.225927
Theta
159.776731
Tajima's D
2.868158
CLR
0.549429
CSRT
0.970201489925504
Interpretation
Uncertain
