Gene
KWMTBOMO06165
Pre Gene Modal
BGIBMGA009638
Annotation
PREDICTED:_uncharacterized_protein_LOC106131099_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.023
Sequence
CDS
ATGTCTTATTTGGGAAATTTACCAATTTACGATCATAAAACATGTGAATGGTCTATTTTCAAGAGTCGTTTAACACAGTTTTTAAAAGTAAATGACGTATCGGAAGAAAAGAAAAGTGCTATCCTCATTACGCACCTCTCCGATGAGGCGTACCGTCTAGTGCGAAATTTAGCATACCCCAAAGATGCTGATTCATTTTCGTATGTGGAGCTGGTTTCATTACTCGATAATCATTTTAAGCCAAAGAAGTGTTCATATACCGACAAGGCTCGTTTTTATGGGGCTACGAGAACCCCAGGCGAATCATTAGGAGACTGGGCGGCGCGACTTAGGGGGTTGGCAAGTTATTGCGAATTCGGCGCGGCTTTGGAAACGAACCTTACGGATCGTTTTGTCCTAGGCCTGGGCTCAGGTCCAGCACGAGATAAGCTGTACGAGCAGGACCCATCAAAAATGAAAATCGATCGAGCGTTAGAAATAGCCCAGCAGGCAGAGAGTGCGAGGGAGGCTAAGTTCATGGAAGTTAGTGTTAAGGAAGAACCTGTGTACCGTGCCTCCTTCACGCTCGACGATAGACGTGAAGCTAATCGACGAGCGCGGACGAGTGAGCCAAGCAGTGGTTATTCAGGGCAGCGGCAGCGGTCCCACCTTGGTGATGGTCAATCAGTTCGCGGGCAGCAACCACGTCGTGACTTCCCAAGGTGCTCGGTTTGCGGGTTAAGAAACCACGACGAAGAGAAGTGCCGTTACAAGGGCTATCGCTGTCAGAAGTGCGGTGAAAAGGGACATTTAAAAAAAGTGTGTGACAAAAATGTGCAGATCAATAATATTTGTGATGACCACGATAAAGGTGTCGGCCACATTCCGGATCCTGACCATTGTGAGGAATGTCATAATTATAGTTTAAAGTACGTGTCTGATAAGCCGATTAATTTAAAATTATTGCTAGGTGGCAAGAGCATAACAATGGAGTTAGATTCTGGTTCTGGGACATCAGTGATATCAAAATCATTATACGATACATTGTTTTCGAAGTACAAGTTAAATAAATGTAACATCAAAATGTGCCTTTATAACGGACATAAAATTTCACCGGAAGGGTATTTTGATATCGAGGCAAAATTTAATCAACAACAAAAAAACTTAAAATTATTTGTTATTAAAAACGGTGGCCCTCCACTTCTCGGTCGAGATTTTATGTCCGCCTTCGGTGTAACGTTTACCACTGGTCTTCATAACATTTGTTATACCGATAAAGATGTTCAAAGTTTGCTAGAACAATTTCCAGATCTGTGGCGTGACGAATTAGGGTCATTCACAAAATTTCAGGTCGATTTAAAATTAAAAGAGGATGCTGTACCAAAGTTTTTCAAACCTCGAACGGTACCATTCGCGCTCAAAGCTAAGGTAGAAGCCGAGTTGGATAGAAATTTTATTCCCAACGCATCATGTATTATGAGTCCACTTCACGAGTTACTCCGCAACGGCGCGCAGTGGGAATGGACGGAGCGGCAACGTCGCGCGGTGCATCAGGTAAAGACCGAGTTGGCATCGGAGCGCGTCCTTGCCCACTTCGAGCACGACGCTCAGCTGATTCTGACCGTAGATGCGGGCCCAGGCGGCCTCGGCGCGGTGCTGGCCCAGAGGCATATCGATCAGATTCTTAGAAGTAAGGAAGATTTTGAAAAAAAGAGTTGTATTACTAGTACGTCGCCTCCATTTTCCAACACAACAATTTTAGCAAAACCTAACAAAACCAATTATAGCGATGGTTCGATTATGACTTCTGTAGCTCAACAGGCTGTGGGAATGGAAAGTGGAGACGGACGTAACGAGCAAGAAAATGTAGTAACCGATATTGGTGACATTGTCCAGCCAACTGACGAGATCTGGGGTGATTGCGAAGAAGTTGACCTTGAACGGGAGAACCTAGTTCCTGGCAGTTCAGCCGAGCCTCCGGCGCCGCCGCTGGCGTTGTCGTCACCAGTCGCGTCTAAACGTACCTCACCTGTTTCTGATCCACATCAGCCCGGACCCTCGTCCTTAACTTATTCCCCACCGCTCACACGTCGCAAGAAGAGAGTTGATTACAAGCAATTCTATTATTGA
Protein
MSYLGNLPIYDHKTCEWSIFKSRLTQFLKVNDVSEEKKSAILITHLSDEAYRLVRNLAYPKDADSFSYVELVSLLDNHFKPKKCSYTDKARFYGATRTPGESLGDWAARLRGLASYCEFGAALETNLTDRFVLGLGSGPARDKLYEQDPSKMKIDRALEIAQQAESAREAKFMEVSVKEEPVYRASFTLDDRREANRRARTSEPSSGYSGQRQRSHLGDGQSVRGQQPRRDFPRCSVCGLRNHDEEKCRYKGYRCQKCGEKGHLKKVCDKNVQINNICDDHDKGVGHIPDPDHCEECHNYSLKYVSDKPINLKLLLGGKSITMELDSGSGTSVISKSLYDTLFSKYKLNKCNIKMCLYNGHKISPEGYFDIEAKFNQQQKNLKLFVIKNGGPPLLGRDFMSAFGVTFTTGLHNICYTDKDVQSLLEQFPDLWRDELGSFTKFQVDLKLKEDAVPKFFKPRTVPFALKAKVEAELDRNFIPNASCIMSPLHELLRNGAQWEWTERQRRAVHQVKTELASERVLAHFEHDAQLILTVDAGPGGLGAVLAQRHIDQILRSKEDFEKKSCITSTSPPFSNTTILAKPNKTNYSDGSIMTSVAQQAVGMESGDGRNEQENVVTDIGDIVQPTDEIWGDCEEVDLERENLVPGSSAEPPAPPLALSSPVASKRTSPVSDPHQPGPSSLTYSPPLTRRKKRVDYKQFYY
Summary
Uniprot
J9KFN3
X1XT86
A0A1Y1KCE7
J9LDN7
X1WTX7
J9KQR5
+ More
A0A146M835 V5GQ27 A0A1Y1LPV1 A0A0A9WT47 J9L142 A0A1Y1NDL2 A0A1Y1NGB8 A0A2R7X643 A0A1B6DD07 A0A1B6CPB9 A0A3B3H2N7 A0A3P8QD94 A0A3P8NHI5 A0A3S2PB62 X1WZX1 A0A3Q1EQC9 A0A3S0ZSV8 A0A1B6E6W0 A0A3P8Q506 A0A1B6CK52 A0A1A7YSF0 A0A1X7TIE8 A0A2I4C6V0 A0A0J7KBM1 A0A1B6KDA3 A0A1B6L0V9
A0A146M835 V5GQ27 A0A1Y1LPV1 A0A0A9WT47 J9L142 A0A1Y1NDL2 A0A1Y1NGB8 A0A2R7X643 A0A1B6DD07 A0A1B6CPB9 A0A3B3H2N7 A0A3P8QD94 A0A3P8NHI5 A0A3S2PB62 X1WZX1 A0A3Q1EQC9 A0A3S0ZSV8 A0A1B6E6W0 A0A3P8Q506 A0A1B6CK52 A0A1A7YSF0 A0A1X7TIE8 A0A2I4C6V0 A0A0J7KBM1 A0A1B6KDA3 A0A1B6L0V9
EMBL
ABLF02036937
ABLF02012266
ABLF02012271
GEZM01092768
GEZM01092767
JAV56467.1
+ More
ABLF02030002 ABLF02008248 ABLF02008249 ABLF02008250 ABLF02035153 ABLF02037855 ABLF02037866 ABLF02061872 ABLF02006499 ABLF02041879 GDHC01002706 JAQ15923.1 GALX01002217 JAB66249.1 GEZM01051399 JAV74971.1 GBHO01031997 JAG11607.1 ABLF02012827 ABLF02012828 ABLF02015552 ABLF02041322 GEZM01005739 JAV95951.1 GEZM01005740 JAV95950.1 KK857512 PTY27246.1 GEDC01013717 JAS23581.1 GEDC01021971 JAS15327.1 RSAL01000130 RVE46453.1 ABLF02013313 RQTK01000185 RUS85038.1 GEDC01007675 GEDC01003661 JAS29623.1 JAS33637.1 GEDC01023506 GEDC01023205 JAS13792.1 JAS14093.1 HADX01010638 SBP32870.1 LBMM01010146 KMQ87646.1 GEBQ01030602 JAT09375.1 GEBQ01022644 GEBQ01012433 JAT17333.1 JAT27544.1
ABLF02030002 ABLF02008248 ABLF02008249 ABLF02008250 ABLF02035153 ABLF02037855 ABLF02037866 ABLF02061872 ABLF02006499 ABLF02041879 GDHC01002706 JAQ15923.1 GALX01002217 JAB66249.1 GEZM01051399 JAV74971.1 GBHO01031997 JAG11607.1 ABLF02012827 ABLF02012828 ABLF02015552 ABLF02041322 GEZM01005739 JAV95951.1 GEZM01005740 JAV95950.1 KK857512 PTY27246.1 GEDC01013717 JAS23581.1 GEDC01021971 JAS15327.1 RSAL01000130 RVE46453.1 ABLF02013313 RQTK01000185 RUS85038.1 GEDC01007675 GEDC01003661 JAS29623.1 JAS33637.1 GEDC01023506 GEDC01023205 JAS13792.1 JAS14093.1 HADX01010638 SBP32870.1 LBMM01010146 KMQ87646.1 GEBQ01030602 JAT09375.1 GEBQ01022644 GEBQ01012433 JAT17333.1 JAT27544.1
Proteomes
Pfam
Interpro
IPR012337
RNaseH-like_sf
+ More
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR000477 RT_dom
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR041373 RT_RNaseH
IPR036875 Znf_CCHC_sf
IPR006612 THAP_Znf
IPR001878 Znf_CCHC
IPR001995 Peptidase_A2_cat
IPR001753 Enoyl-CoA_hydra/iso
IPR029045 ClpP/crotonase-like_dom_sf
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR000477 RT_dom
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR041373 RT_RNaseH
IPR036875 Znf_CCHC_sf
IPR006612 THAP_Znf
IPR001878 Znf_CCHC
IPR001995 Peptidase_A2_cat
IPR001753 Enoyl-CoA_hydra/iso
IPR029045 ClpP/crotonase-like_dom_sf
Gene 3D
ProteinModelPortal
J9KFN3
X1XT86
A0A1Y1KCE7
J9LDN7
X1WTX7
J9KQR5
+ More
A0A146M835 V5GQ27 A0A1Y1LPV1 A0A0A9WT47 J9L142 A0A1Y1NDL2 A0A1Y1NGB8 A0A2R7X643 A0A1B6DD07 A0A1B6CPB9 A0A3B3H2N7 A0A3P8QD94 A0A3P8NHI5 A0A3S2PB62 X1WZX1 A0A3Q1EQC9 A0A3S0ZSV8 A0A1B6E6W0 A0A3P8Q506 A0A1B6CK52 A0A1A7YSF0 A0A1X7TIE8 A0A2I4C6V0 A0A0J7KBM1 A0A1B6KDA3 A0A1B6L0V9
A0A146M835 V5GQ27 A0A1Y1LPV1 A0A0A9WT47 J9L142 A0A1Y1NDL2 A0A1Y1NGB8 A0A2R7X643 A0A1B6DD07 A0A1B6CPB9 A0A3B3H2N7 A0A3P8QD94 A0A3P8NHI5 A0A3S2PB62 X1WZX1 A0A3Q1EQC9 A0A3S0ZSV8 A0A1B6E6W0 A0A3P8Q506 A0A1B6CK52 A0A1A7YSF0 A0A1X7TIE8 A0A2I4C6V0 A0A0J7KBM1 A0A1B6KDA3 A0A1B6L0V9
PDB
4OL8
E-value=9.21836e-08,
Score=137
Ontologies
GO
Topology
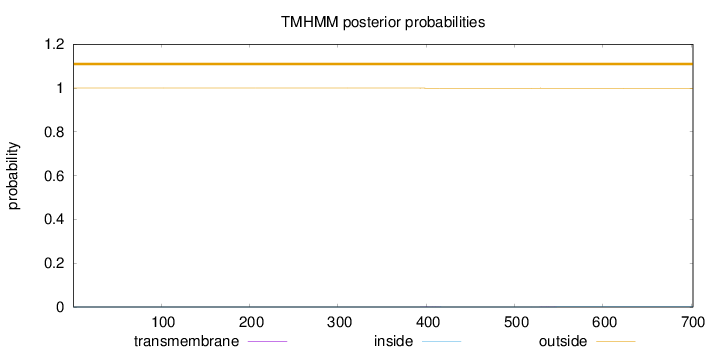
Length:
702
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01239
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00008
outside
1 - 702
Population Genetic Test Statistics
Pi
243.139591
Theta
162.682723
Tajima's D
1.395871
CLR
10.814854
CSRT
0.764361781910904
Interpretation
Uncertain
