Gene
KWMTBOMO06149 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001754
Annotation
PREDICTED:_gelsolin-like_[Amyelois_transitella]
Full name
Gelsolin
Location in the cell
Cytoplasmic Reliability : 2.27
Sequence
CDS
ATGAAAGCCATCACCGTAGCCAACCAGATTAAAGACCAGGATCACAATGGAAGGGCTGATATCGAAGTGGTCGACTCCGAAGCGCACGATGGAATCTTCGACAGGTTCTTCGATGCCCTCGGCTCCGGCAACAAGGAGGCCATCGCTGATGCCGAAGAAGGGGGCGACGATCAAGAGTTCGAACGCGTCGTCACGAAGGAGGTGTCCCTGAGTGAGATCTCCGATAAGACTGGCAGCATCAGGATAACGAGACTCCCCGCGCCACTTAAGAAGGAGCAACTGAACACCGATGAATGCTACGTGCTTGATACCGGAAGTTACAGCGGCATCTATGTGTGGGTCGGGCGCGGCTCGAACCTAAAGGAGAAAGCCGAAGCGATGAATAGAGCTGAGCAATACCTGAAATACCACGACTATCCGGCTTGGGTCCACGTGTCGAAGGTTCCGGAAGGCGCTGAACCGATACCCTTCAAGCAATACTTCGAGGATTGGAACTGA
Protein
MKAITVANQIKDQDHNGRADIEVVDSEAHDGIFDRFFDALGSGNKEAIADAEEGGDDQEFERVVTKEVSLSEISDKTGSIRITRLPAPLKKEQLNTDECYVLDTGSYSGIYVWVGRGSNLKEKAEAMNRAEQYLKYHDYPAWVHVSKVPEGAEPIPFKQYFEDWN
Summary
Description
Calcium-regulated, actin-modulating protein that binds to the plus (or barbed) ends of actin monomers or filaments, preventing monomer exchange (end-blocking or capping). It can promote the assembly of monomers into filaments (nucleation) as well as sever filaments already formed.
Subunit
Binds to actin and to fibronectin.
Similarity
Belongs to the villin/gelsolin family.
Keywords
Actin-binding
Alternative splicing
Calcium
Complete proteome
Cytoplasm
Cytoskeleton
Metal-binding
Phosphoprotein
Reference proteome
Repeat
Secreted
Signal
Feature
chain Gelsolin
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9IWX1
A0A2W1BN17
A0A2H1WKP3
A0A0N0PA91
A0A2A4JQW7
A0A2H1X307
+ More
A0A2A4JSL9 A0A0N1PF90 I4DKR3 A0A0N1I6N5 E5L9L1 A0A2K8JM62 T1HKR8 A0A1S4E792 B4NFJ3 A0A224XF20 A0A0M3QXF2 A0A161MIR2 Q07171-2 A0A0C4DHG6 A0A0C4DHG2 Q07171 A0A224XQJ4 Q07171-6 A0A1A9WIF6 B4QUY4 A0A1I8NCV8 T1PDD6 B4JS24 A0A069DYQ6 I5APK8 A0A1I8NXE8 A0A1I8NXC0 Q295D4 B4GM71 A0A0Q9XFV1 A0A0Q9XDT7 B4KCW4 A0A1W4VZL6 A0A0P4VVN5 A0A0R1DZS5 A0A3B0K219 B4PTP4 A0A1W4VLN5 A0A0N8P1H5 A0A0Q9WBU6 A0A171A3J0 B4MBI4 B3LXY4 A0A0K8VYB1 A0A336MRC3 A0A1B0FCE4 A0A336MY83 U5ETK7 A0A0A1X561 A0A1J1HKI1 A0A0K8WEL2 A0A2K8JW66 D6W686 A0A034VAZ1 A0A0T6ATX5 J9K5M7 A0A034V9E0 A0A293LI79 A0A0A1XCB7 A0A0K8SZE6 A0A2R5L4E0 A0A2S2PJ12 A0A2H8U180 W8BFB5 A0A1L8E5G4 A0A1L8E5M0 A0A1L8E606 A0A1L8E5B4 A0A1L8E5D0 A0A1L8E5N5 A0A0K8WH15 A0A0K8VEK4 B3P1X4 A0A336MF08 L7MCZ6 A0A224YQ72 A0A1E1XM95 A0A224YNC3 A0A1B0BZ83 A0A1Z5L535 A0A034VNT3 A0A034VMT8 B7PVR6 A0A131XHX0 A0A131YKS1 A0A1E1XAG7 A0A0L0CJJ3 W8B6S7 A0A1E1W458 A0A0A9WEB4 V5HJ32 A0A2J7RA32 A0A1A9UQ65 A0A1B6CL01
A0A2A4JSL9 A0A0N1PF90 I4DKR3 A0A0N1I6N5 E5L9L1 A0A2K8JM62 T1HKR8 A0A1S4E792 B4NFJ3 A0A224XF20 A0A0M3QXF2 A0A161MIR2 Q07171-2 A0A0C4DHG6 A0A0C4DHG2 Q07171 A0A224XQJ4 Q07171-6 A0A1A9WIF6 B4QUY4 A0A1I8NCV8 T1PDD6 B4JS24 A0A069DYQ6 I5APK8 A0A1I8NXE8 A0A1I8NXC0 Q295D4 B4GM71 A0A0Q9XFV1 A0A0Q9XDT7 B4KCW4 A0A1W4VZL6 A0A0P4VVN5 A0A0R1DZS5 A0A3B0K219 B4PTP4 A0A1W4VLN5 A0A0N8P1H5 A0A0Q9WBU6 A0A171A3J0 B4MBI4 B3LXY4 A0A0K8VYB1 A0A336MRC3 A0A1B0FCE4 A0A336MY83 U5ETK7 A0A0A1X561 A0A1J1HKI1 A0A0K8WEL2 A0A2K8JW66 D6W686 A0A034VAZ1 A0A0T6ATX5 J9K5M7 A0A034V9E0 A0A293LI79 A0A0A1XCB7 A0A0K8SZE6 A0A2R5L4E0 A0A2S2PJ12 A0A2H8U180 W8BFB5 A0A1L8E5G4 A0A1L8E5M0 A0A1L8E606 A0A1L8E5B4 A0A1L8E5D0 A0A1L8E5N5 A0A0K8WH15 A0A0K8VEK4 B3P1X4 A0A336MF08 L7MCZ6 A0A224YQ72 A0A1E1XM95 A0A224YNC3 A0A1B0BZ83 A0A1Z5L535 A0A034VNT3 A0A034VMT8 B7PVR6 A0A131XHX0 A0A131YKS1 A0A1E1XAG7 A0A0L0CJJ3 W8B6S7 A0A1E1W458 A0A0A9WEB4 V5HJ32 A0A2J7RA32 A0A1A9UQ65 A0A1B6CL01
Pubmed
19121390
28756777
26354079
22651552
20978910
17994087
+ More
8386771 8175883 10731132 12537572 12537569 12537568 12537573 12537574 16110336 17569856 17569867 25315136 26334808 15632085 18057021 27129103 17550304 25830018 18362917 19820115 25348373 24495485 25576852 28797301 29209593 28528879 28049606 26830274 28503490 26108605 25401762 25765539
8386771 8175883 10731132 12537572 12537569 12537568 12537573 12537574 16110336 17569856 17569867 25315136 26334808 15632085 18057021 27129103 17550304 25830018 18362917 19820115 25348373 24495485 25576852 28797301 29209593 28528879 28049606 26830274 28503490 26108605 25401762 25765539
EMBL
BABH01021068
KZ149957
PZC76449.1
ODYU01009215
SOQ53482.1
KQ458725
+ More
KPJ05340.1 NWSH01000756 PCG74391.1 ODYU01013020 SOQ59632.1 PCG74392.1 KPJ05342.1 AK401881 BAM18503.1 KQ460736 KPJ12596.1 HQ383992 ADQ64485.1 KY030980 ATU82731.1 ACPB03022068 CH964251 EDW83060.2 GFTR01005349 JAW11077.1 CP012526 ALC45823.1 GEMB01001542 JAS01616.1 L08794 X75629 X75630 AE014297 AY119662 BT031288 AAF52163.1 ABI31119.2 AAO41509.3 GFTR01005706 JAW10720.1 CM000364 EDX11565.1 KA646135 AFP60764.1 CH916373 EDV94564.1 GBGD01001975 JAC86914.1 CM000070 EIM52893.1 EAL28778.1 CH479185 EDW37945.1 CH933806 KRG02435.1 KRG02433.1 EDW16986.1 KRG02434.1 GDKW01002904 JAI53691.1 CM000160 KRK02572.1 OUUW01000014 SPP88317.1 EDW95627.1 CH902617 KPU79940.1 CH940656 KRF78299.1 GEMB01001541 JAS01617.1 EDW58455.1 EDV42840.1 GDHF01008411 JAI43903.1 UFQT01001498 SSX30817.1 CCAG010002582 CCAG010002583 SSX30818.1 GANO01002735 JAB57136.1 GBXI01017400 GBXI01008256 JAC96891.1 JAD06036.1 CVRI01000008 CRK88567.1 GDHF01002830 JAI49484.1 MF683379 ATU82520.1 KQ971306 EFA11080.2 GAKP01020259 JAC38693.1 LJIG01022826 KRT78520.1 ABLF02022767 GAKP01020261 GAKP01020260 JAC38692.1 GFWV01008468 MAA33197.1 GBXI01005705 JAD08587.1 GBRD01007158 JAG58663.1 GGLE01000184 MBY04310.1 GGMR01016820 MBY29439.1 GFXV01007996 MBW19801.1 GAMC01006570 JAB99985.1 GFDF01000141 JAV13943.1 GFDF01000140 JAV13944.1 GFDF01000142 JAV13942.1 GFDF01000144 JAV13940.1 GFDF01000143 JAV13941.1 GFDF01000145 JAV13939.1 GDHF01032030 GDHF01001868 JAI20284.1 JAI50446.1 GDHF01015067 GDHF01010278 JAI37247.1 JAI42036.1 CH954181 EDV47747.1 UFQS01001020 UFQT01001020 SSX08604.1 SSX28520.1 GACK01002948 JAA62086.1 GFPF01007949 MAA19095.1 GFAA01003228 JAU00207.1 GFPF01007950 MAA19096.1 JXJN01023019 JXJN01023020 GFJQ02004488 JAW02482.1 GAKP01015191 GAKP01015190 JAC43762.1 GAKP01015189 GAKP01015188 JAC43764.1 ABJB010273219 ABJB010277958 ABJB010343362 ABJB010707454 ABJB011053052 DS801915 EEC10688.1 GEFH01002776 JAP65805.1 GEDV01009010 JAP79547.1 GFAC01002976 JAT96212.1 JRES01000310 KNC32416.1 GAMC01012223 JAB94332.1 GDQN01009286 JAT81768.1 GBHO01036802 JAG06802.1 GANP01006869 JAB77599.1 NEVH01006578 PNF37693.1 GEDC01023154 JAS14144.1
KPJ05340.1 NWSH01000756 PCG74391.1 ODYU01013020 SOQ59632.1 PCG74392.1 KPJ05342.1 AK401881 BAM18503.1 KQ460736 KPJ12596.1 HQ383992 ADQ64485.1 KY030980 ATU82731.1 ACPB03022068 CH964251 EDW83060.2 GFTR01005349 JAW11077.1 CP012526 ALC45823.1 GEMB01001542 JAS01616.1 L08794 X75629 X75630 AE014297 AY119662 BT031288 AAF52163.1 ABI31119.2 AAO41509.3 GFTR01005706 JAW10720.1 CM000364 EDX11565.1 KA646135 AFP60764.1 CH916373 EDV94564.1 GBGD01001975 JAC86914.1 CM000070 EIM52893.1 EAL28778.1 CH479185 EDW37945.1 CH933806 KRG02435.1 KRG02433.1 EDW16986.1 KRG02434.1 GDKW01002904 JAI53691.1 CM000160 KRK02572.1 OUUW01000014 SPP88317.1 EDW95627.1 CH902617 KPU79940.1 CH940656 KRF78299.1 GEMB01001541 JAS01617.1 EDW58455.1 EDV42840.1 GDHF01008411 JAI43903.1 UFQT01001498 SSX30817.1 CCAG010002582 CCAG010002583 SSX30818.1 GANO01002735 JAB57136.1 GBXI01017400 GBXI01008256 JAC96891.1 JAD06036.1 CVRI01000008 CRK88567.1 GDHF01002830 JAI49484.1 MF683379 ATU82520.1 KQ971306 EFA11080.2 GAKP01020259 JAC38693.1 LJIG01022826 KRT78520.1 ABLF02022767 GAKP01020261 GAKP01020260 JAC38692.1 GFWV01008468 MAA33197.1 GBXI01005705 JAD08587.1 GBRD01007158 JAG58663.1 GGLE01000184 MBY04310.1 GGMR01016820 MBY29439.1 GFXV01007996 MBW19801.1 GAMC01006570 JAB99985.1 GFDF01000141 JAV13943.1 GFDF01000140 JAV13944.1 GFDF01000142 JAV13942.1 GFDF01000144 JAV13940.1 GFDF01000143 JAV13941.1 GFDF01000145 JAV13939.1 GDHF01032030 GDHF01001868 JAI20284.1 JAI50446.1 GDHF01015067 GDHF01010278 JAI37247.1 JAI42036.1 CH954181 EDV47747.1 UFQS01001020 UFQT01001020 SSX08604.1 SSX28520.1 GACK01002948 JAA62086.1 GFPF01007949 MAA19095.1 GFAA01003228 JAU00207.1 GFPF01007950 MAA19096.1 JXJN01023019 JXJN01023020 GFJQ02004488 JAW02482.1 GAKP01015191 GAKP01015190 JAC43762.1 GAKP01015189 GAKP01015188 JAC43764.1 ABJB010273219 ABJB010277958 ABJB010343362 ABJB010707454 ABJB011053052 DS801915 EEC10688.1 GEFH01002776 JAP65805.1 GEDV01009010 JAP79547.1 GFAC01002976 JAT96212.1 JRES01000310 KNC32416.1 GAMC01012223 JAB94332.1 GDQN01009286 JAT81768.1 GBHO01036802 JAG06802.1 GANP01006869 JAB77599.1 NEVH01006578 PNF37693.1 GEDC01023154 JAS14144.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000015103
UP000079169
+ More
UP000007798 UP000092553 UP000000803 UP000091820 UP000000304 UP000095301 UP000001070 UP000001819 UP000095300 UP000008744 UP000009192 UP000192221 UP000002282 UP000268350 UP000007801 UP000008792 UP000092444 UP000183832 UP000007266 UP000007819 UP000008711 UP000092460 UP000001555 UP000037069 UP000235965 UP000078200
UP000007798 UP000092553 UP000000803 UP000091820 UP000000304 UP000095301 UP000001070 UP000001819 UP000095300 UP000008744 UP000009192 UP000192221 UP000002282 UP000268350 UP000007801 UP000008792 UP000092444 UP000183832 UP000007266 UP000007819 UP000008711 UP000092460 UP000001555 UP000037069 UP000235965 UP000078200
Interpro
Gene 3D
ProteinModelPortal
H9IWX1
A0A2W1BN17
A0A2H1WKP3
A0A0N0PA91
A0A2A4JQW7
A0A2H1X307
+ More
A0A2A4JSL9 A0A0N1PF90 I4DKR3 A0A0N1I6N5 E5L9L1 A0A2K8JM62 T1HKR8 A0A1S4E792 B4NFJ3 A0A224XF20 A0A0M3QXF2 A0A161MIR2 Q07171-2 A0A0C4DHG6 A0A0C4DHG2 Q07171 A0A224XQJ4 Q07171-6 A0A1A9WIF6 B4QUY4 A0A1I8NCV8 T1PDD6 B4JS24 A0A069DYQ6 I5APK8 A0A1I8NXE8 A0A1I8NXC0 Q295D4 B4GM71 A0A0Q9XFV1 A0A0Q9XDT7 B4KCW4 A0A1W4VZL6 A0A0P4VVN5 A0A0R1DZS5 A0A3B0K219 B4PTP4 A0A1W4VLN5 A0A0N8P1H5 A0A0Q9WBU6 A0A171A3J0 B4MBI4 B3LXY4 A0A0K8VYB1 A0A336MRC3 A0A1B0FCE4 A0A336MY83 U5ETK7 A0A0A1X561 A0A1J1HKI1 A0A0K8WEL2 A0A2K8JW66 D6W686 A0A034VAZ1 A0A0T6ATX5 J9K5M7 A0A034V9E0 A0A293LI79 A0A0A1XCB7 A0A0K8SZE6 A0A2R5L4E0 A0A2S2PJ12 A0A2H8U180 W8BFB5 A0A1L8E5G4 A0A1L8E5M0 A0A1L8E606 A0A1L8E5B4 A0A1L8E5D0 A0A1L8E5N5 A0A0K8WH15 A0A0K8VEK4 B3P1X4 A0A336MF08 L7MCZ6 A0A224YQ72 A0A1E1XM95 A0A224YNC3 A0A1B0BZ83 A0A1Z5L535 A0A034VNT3 A0A034VMT8 B7PVR6 A0A131XHX0 A0A131YKS1 A0A1E1XAG7 A0A0L0CJJ3 W8B6S7 A0A1E1W458 A0A0A9WEB4 V5HJ32 A0A2J7RA32 A0A1A9UQ65 A0A1B6CL01
A0A2A4JSL9 A0A0N1PF90 I4DKR3 A0A0N1I6N5 E5L9L1 A0A2K8JM62 T1HKR8 A0A1S4E792 B4NFJ3 A0A224XF20 A0A0M3QXF2 A0A161MIR2 Q07171-2 A0A0C4DHG6 A0A0C4DHG2 Q07171 A0A224XQJ4 Q07171-6 A0A1A9WIF6 B4QUY4 A0A1I8NCV8 T1PDD6 B4JS24 A0A069DYQ6 I5APK8 A0A1I8NXE8 A0A1I8NXC0 Q295D4 B4GM71 A0A0Q9XFV1 A0A0Q9XDT7 B4KCW4 A0A1W4VZL6 A0A0P4VVN5 A0A0R1DZS5 A0A3B0K219 B4PTP4 A0A1W4VLN5 A0A0N8P1H5 A0A0Q9WBU6 A0A171A3J0 B4MBI4 B3LXY4 A0A0K8VYB1 A0A336MRC3 A0A1B0FCE4 A0A336MY83 U5ETK7 A0A0A1X561 A0A1J1HKI1 A0A0K8WEL2 A0A2K8JW66 D6W686 A0A034VAZ1 A0A0T6ATX5 J9K5M7 A0A034V9E0 A0A293LI79 A0A0A1XCB7 A0A0K8SZE6 A0A2R5L4E0 A0A2S2PJ12 A0A2H8U180 W8BFB5 A0A1L8E5G4 A0A1L8E5M0 A0A1L8E606 A0A1L8E5B4 A0A1L8E5D0 A0A1L8E5N5 A0A0K8WH15 A0A0K8VEK4 B3P1X4 A0A336MF08 L7MCZ6 A0A224YQ72 A0A1E1XM95 A0A224YNC3 A0A1B0BZ83 A0A1Z5L535 A0A034VNT3 A0A034VMT8 B7PVR6 A0A131XHX0 A0A131YKS1 A0A1E1XAG7 A0A0L0CJJ3 W8B6S7 A0A1E1W458 A0A0A9WEB4 V5HJ32 A0A2J7RA32 A0A1A9UQ65 A0A1B6CL01
PDB
1J72
E-value=1.46472e-15,
Score=197
Ontologies
GO
PANTHER
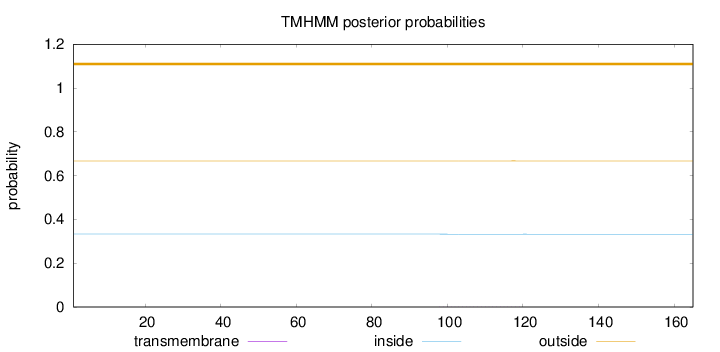
Topology
Subcellular location
Secreted
Length:
165
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01131
Exp number, first 60 AAs:
0
Total prob of N-in:
0.33296
outside
1 - 165
Population Genetic Test Statistics
Pi
23.525135
Theta
148.975249
Tajima's D
1.625539
CLR
0.248001
CSRT
0.814309284535773
Interpretation
Uncertain
