Gene
KWMTBOMO06144
Pre Gene Modal
BGIBMGA001750
Annotation
FKBP6_BOMMO_RecName:_Full%3DInactive_peptidyl-prolyl_cis-trans_isomerase_shutdown
Full name
Inactive peptidyl-prolyl cis-trans isomerase shutdown
+ More
Peptidylprolyl isomerase
Peptidylprolyl isomerase
Location in the cell
Cytoplasmic Reliability : 3.938
Sequence
CDS
ATGGAGCAAGTATTTCAAGAACCCGTTCAACTGTCTAAAGGAATAAATCTGAGAGAGCTCACTTCAGAAATGGAGTTTCAGATTGATGTGGACTTTCAAAACAGAAAAACGGAGATGTTCATCAACGGTGATGACGATTTGTTTCCTGAAATGGGTGACGAGGATGACAGTGATGGAGAGGATACACTCAGGAATATTGAGGAGACAACCAAAAAGATGATGCTGGCCTCACCAGATTACTTGTCTTTTGAGGAGTTGGGCGCCAATATGATAGACTGCATGAGCACTGGTGACGTGAAAATGCTCATCATAGAAGAAGGTAAAGGACCTCTGGTGCCCGTTGATTCAGAAGTGACGATACATTACGCGGCTTACTGGGAGAAAGCTGTGATACCGTTTGATTCTACTTTAACCATGAATATGGGCGCACCAAAGCGTCTGCGTTTAGGAACAGGGAAAATAATACCCGGTCTGGAGATAGGTTTGACTATGGTCAAAGGGCCACAGGCGCGTCTCAACTTGCTGGTGCAACCGGCGGCCGCGTGGGGTCCGCGCGGGGTCCCGCCCAGGATCAGACCGGAGCCCGCGCTCTTCGTCATCGTCTTGTATGACGTCAAAGACAACTACGCGGCCACCAGATTTAACGATCTACCAATGGCGGAGCAGACTAAATACGAGGTCACATTGCGTACCGTCAACGCCTTACGCGCTGACGCCAAAGAGCTATACAAGAAAAAGAAATATGTGAAGGCCATTAAGAATTATCAGCAGGCCATCTCGGTTTTGCGTCTATCGCGTCCTGGTACGGCCGACGAGGAGTTAGACATAAAAAACAATAAAGTTAACGCGTACTTAAATTTGGCAGTCTGCTATTACAAGACGAACAAGCCTAAACACGTTCTGAACATGTGCGAATGCTTGGACAGACTTATTGACACCGAAAAGCACTGCAAAGCGCTTTACTACTATGGGAAAGCTCACGAAATGCTCGGCAAAACGGAAATGGCGATTAAATATTACAAGAAAGCTTTAAAATTGGAGCCGAAAAATAAAGAGATTGGCAAGATATTGGCGGATTTAGATACGAAGACGAAGGATTTTGCGGTCAACGAGAAGGCTATGTGGCTGAAGGCGTTTAACGTCGAGGTCCCCGCGACGAACGCGGTCTATGACGTTGACGCGACTTTCCAAAGTGACGTGTTGGACATGTGTCAAAGTTTGGCGGGAAGGTCGGAGTTTGCGAAATTCGATCTGCCCGTCGGGCTGACGAAAAACGAGGTGGATTGCATCAAGGGTATTGTGAGCGATTTCGGGTGCCTCTCAGTGGACGTCAGCGGTGAAGGTAGAAATAAAAAAGTTTGTATCGTTAAGAAGATCGTCTGA
Protein
MEQVFQEPVQLSKGINLRELTSEMEFQIDVDFQNRKTEMFINGDDDLFPEMGDEDDSDGEDTLRNIEETTKKMMLASPDYLSFEELGANMIDCMSTGDVKMLIIEEGKGPLVPVDSEVTIHYAAYWEKAVIPFDSTLTMNMGAPKRLRLGTGKIIPGLEIGLTMVKGPQARLNLLVQPAAAWGPRGVPPRIRPEPALFVIVLYDVKDNYAATRFNDLPMAEQTKYEVTLRTVNALRADAKELYKKKKYVKAIKNYQQAISVLRLSRPGTADEELDIKNNKVNAYLNLAVCYYKTNKPKHVLNMCECLDRLIDTEKHCKALYYYGKAHEMLGKTEMAIKYYKKALKLEPKNKEIGKILADLDTKTKDFAVNEKAMWLKAFNVEVPATNAVYDVDATFQSDVLDMCQSLAGRSEFAKFDLPVGLTKNEVDCIKGIVSDFGCLSVDVSGEGRNKKVCIVKKIV
Summary
Description
Co-chaperone required during oogenesis to repress transposable elements and prevent their mobilization, which is essential for the germline integrity. Acts via the piRNA metabolic process, which mediates the repression of transposable elements during meiosis by forming complexes composed of piRNAs and Piwi proteins and govern the methylation and subsequent repression of transposons. Acts as a co-chaperone via its interaction with Hsp83/HSP90 and is required for the biogenesis of all three piRNA major populations (By similarity).
Catalytic Activity
[protein]-peptidylproline (omega=180) = [protein]-peptidylproline (omega=0)
Subunit
Interacts with Hsp83.
Similarity
Belongs to the FKBP6 family.
Belongs to the AB hydrolase superfamily. Lipase family.
Belongs to the AB hydrolase superfamily. Lipase family.
Keywords
Complete proteome
Cytoplasm
Differentiation
Meiosis
Oogenesis
Reference proteome
Repeat
RNA-mediated gene silencing
TPR repeat
Feature
chain Inactive peptidyl-prolyl cis-trans isomerase shutdown
Uniprot
H9IWW7
A0A2W1B4N3
A0A0N1I7G7
A0A2A4JPU8
A0A0L7L198
E2A4Y8
+ More
A0A3L8DXT2 A0A026X467 A0A1Y9H221 A0A1X7V8C2 A0A310SIH5 A0A0L0C9J3 T1PHG5 A0A1I8NAG1 F4WY20 A0A0A1XP56 A0A154PEE0 A0A195AYQ9 A0A158NYL1 A0A195DEB8 A0A0J7L1A3 F4X2D5 A0A151WU13 A0A0L7QY84 A0A1B0BNJ8 A0A1A9Y0M9 A0A195FTK4 A0A0K8WCQ6 W8CDP5 A0A0M9A1A3 A0A1A9WIQ6 A0A1I8NY75 A0A2A3EFJ9 A0A034WCL6 A0A182Q1X1 A0A151IEV6 A0A182G8A7 A0A1Q3F193 A0A084VZE6 A0A1A9ZWD6 A0A1B0G7Q4 A0A240PJX4 A0A182R8Q0 A0A182VMP2 E9JDF4 Q16PH6 A0A1S4FTS0 A0A182WF59 A0A1A9V8D6 B0WGW1 U5ENS3 W5JRX6 A0A182JWF1 A0A084VV77 A0A182XCA1 T2MIQ4 A0A182LPI6 A0A182FTD7 E2C0E2 A0A182MGT8 A0A182P9C6 A0A182HNK2 A0A182TN62 A0NFE6 A0A084VU23 A0A2M4BRI7 A0A182T071 A0A146LVU3 A0A182YPF5 B4PA75 A0A194R279 A0A0A9YHD0 A0A0P4W9F9 A0A2L2YF89 A0A1B6F1X8 A0A2B4RN23 A0A023GMQ0 A0A336LDM3 A0A336L5J2 A0A069DYT9
A0A3L8DXT2 A0A026X467 A0A1Y9H221 A0A1X7V8C2 A0A310SIH5 A0A0L0C9J3 T1PHG5 A0A1I8NAG1 F4WY20 A0A0A1XP56 A0A154PEE0 A0A195AYQ9 A0A158NYL1 A0A195DEB8 A0A0J7L1A3 F4X2D5 A0A151WU13 A0A0L7QY84 A0A1B0BNJ8 A0A1A9Y0M9 A0A195FTK4 A0A0K8WCQ6 W8CDP5 A0A0M9A1A3 A0A1A9WIQ6 A0A1I8NY75 A0A2A3EFJ9 A0A034WCL6 A0A182Q1X1 A0A151IEV6 A0A182G8A7 A0A1Q3F193 A0A084VZE6 A0A1A9ZWD6 A0A1B0G7Q4 A0A240PJX4 A0A182R8Q0 A0A182VMP2 E9JDF4 Q16PH6 A0A1S4FTS0 A0A182WF59 A0A1A9V8D6 B0WGW1 U5ENS3 W5JRX6 A0A182JWF1 A0A084VV77 A0A182XCA1 T2MIQ4 A0A182LPI6 A0A182FTD7 E2C0E2 A0A182MGT8 A0A182P9C6 A0A182HNK2 A0A182TN62 A0NFE6 A0A084VU23 A0A2M4BRI7 A0A182T071 A0A146LVU3 A0A182YPF5 B4PA75 A0A194R279 A0A0A9YHD0 A0A0P4W9F9 A0A2L2YF89 A0A1B6F1X8 A0A2B4RN23 A0A023GMQ0 A0A336LDM3 A0A336L5J2 A0A069DYT9
EC Number
5.2.1.8
Pubmed
EMBL
KZ150331
PZC71352.1
KQ460596
KPJ13908.1
NWSH01000868
PCG73786.1
+ More
JTDY01003724 KOB69091.1 GL436770 EFN71505.1 QOIP01000003 RLU25102.1 KK107013 EZA62898.1 KQ759870 OAD62431.1 JRES01000819 KNC28119.1 KA648211 AFP62840.1 GL888434 EGI60945.1 GBXI01001622 JAD12670.1 KQ434875 KZC09784.1 KQ976701 KYM77373.1 ADTU01004020 KQ980953 KYN11177.1 LBMM01001374 KMQ96426.1 GL888577 EGI59395.1 KQ982748 KYQ51324.1 KQ414697 KOC63519.1 JXJN01017416 KQ981276 KYN43793.1 GDHF01003679 GDHF01002901 JAI48635.1 JAI49413.1 GAMC01001586 JAC04970.1 KQ435765 KOX75315.1 KZ288269 PBC29989.1 GAKP01005651 JAC53301.1 AXCN02002114 KQ977854 KYM99352.1 JXUM01151776 KQ571296 KXJ68208.1 GFDL01013731 JAV21314.1 ATLV01018748 KE525248 KFB43340.1 CCAG010016890 GL771866 EFZ09302.1 CH477781 EAT36272.1 DS231930 EDS27255.1 GANO01003957 JAB55914.1 ADMH02000540 ETN66048.1 ATLV01017157 KE525157 KFB41871.1 HAAD01005570 CDG71802.1 GL451770 EFN78601.1 AXCM01000254 APCN01001938 AAAB01008966 EAU76262.2 ATLV01016628 KE525098 KFB41467.1 GGFJ01006491 MBW55632.1 GDHC01006821 JAQ11808.1 CM000158 EDW92401.1 KQ460855 KPJ11893.1 GBHO01014659 GBRD01017776 JAG28945.1 JAG48051.1 GDRN01070793 GDRN01070791 JAI63809.1 IAAA01022285 IAAA01022286 LAA06687.1 GECZ01025595 JAS44174.1 LSMT01000381 PFX19011.1 GBBM01001273 JAC34145.1 UFQS01003771 UFQT01003771 SSX15928.1 SSX35270.1 UFQS01001911 UFQT01001911 SSX12694.1 SSX32137.1 GBGD01001810 JAC87079.1
JTDY01003724 KOB69091.1 GL436770 EFN71505.1 QOIP01000003 RLU25102.1 KK107013 EZA62898.1 KQ759870 OAD62431.1 JRES01000819 KNC28119.1 KA648211 AFP62840.1 GL888434 EGI60945.1 GBXI01001622 JAD12670.1 KQ434875 KZC09784.1 KQ976701 KYM77373.1 ADTU01004020 KQ980953 KYN11177.1 LBMM01001374 KMQ96426.1 GL888577 EGI59395.1 KQ982748 KYQ51324.1 KQ414697 KOC63519.1 JXJN01017416 KQ981276 KYN43793.1 GDHF01003679 GDHF01002901 JAI48635.1 JAI49413.1 GAMC01001586 JAC04970.1 KQ435765 KOX75315.1 KZ288269 PBC29989.1 GAKP01005651 JAC53301.1 AXCN02002114 KQ977854 KYM99352.1 JXUM01151776 KQ571296 KXJ68208.1 GFDL01013731 JAV21314.1 ATLV01018748 KE525248 KFB43340.1 CCAG010016890 GL771866 EFZ09302.1 CH477781 EAT36272.1 DS231930 EDS27255.1 GANO01003957 JAB55914.1 ADMH02000540 ETN66048.1 ATLV01017157 KE525157 KFB41871.1 HAAD01005570 CDG71802.1 GL451770 EFN78601.1 AXCM01000254 APCN01001938 AAAB01008966 EAU76262.2 ATLV01016628 KE525098 KFB41467.1 GGFJ01006491 MBW55632.1 GDHC01006821 JAQ11808.1 CM000158 EDW92401.1 KQ460855 KPJ11893.1 GBHO01014659 GBRD01017776 JAG28945.1 JAG48051.1 GDRN01070793 GDRN01070791 JAI63809.1 IAAA01022285 IAAA01022286 LAA06687.1 GECZ01025595 JAS44174.1 LSMT01000381 PFX19011.1 GBBM01001273 JAC34145.1 UFQS01003771 UFQT01003771 SSX15928.1 SSX35270.1 UFQS01001911 UFQT01001911 SSX12694.1 SSX32137.1 GBGD01001810 JAC87079.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000037510
UP000000311
UP000279307
+ More
UP000053097 UP000075884 UP000007879 UP000037069 UP000095301 UP000007755 UP000076502 UP000078540 UP000005205 UP000078492 UP000036403 UP000075809 UP000053825 UP000092460 UP000092443 UP000078541 UP000053105 UP000091820 UP000095300 UP000242457 UP000075886 UP000078542 UP000069940 UP000249989 UP000030765 UP000092445 UP000092444 UP000075880 UP000075900 UP000075903 UP000008820 UP000075920 UP000078200 UP000002320 UP000000673 UP000075881 UP000076407 UP000075882 UP000069272 UP000008237 UP000075883 UP000075885 UP000075840 UP000075902 UP000007062 UP000075901 UP000076408 UP000002282 UP000225706
UP000053097 UP000075884 UP000007879 UP000037069 UP000095301 UP000007755 UP000076502 UP000078540 UP000005205 UP000078492 UP000036403 UP000075809 UP000053825 UP000092460 UP000092443 UP000078541 UP000053105 UP000091820 UP000095300 UP000242457 UP000075886 UP000078542 UP000069940 UP000249989 UP000030765 UP000092445 UP000092444 UP000075880 UP000075900 UP000075903 UP000008820 UP000075920 UP000078200 UP000002320 UP000000673 UP000075881 UP000076407 UP000075882 UP000069272 UP000008237 UP000075883 UP000075885 UP000075840 UP000075902 UP000007062 UP000075901 UP000076408 UP000002282 UP000225706
PRIDE
Pfam
Interpro
IPR019734
TPR_repeat
+ More
IPR011990 TPR-like_helical_dom_sf
IPR013026 TPR-contain_dom
IPR042282 FKBP6/shu
IPR013105 TPR_2
IPR001179 PPIase_FKBP_dom
IPR002048 EF_hand_dom
IPR011992 EF-hand-dom_pair
IPR018247 EF_Hand_1_Ca_BS
IPR000734 TAG_lipase
IPR029058 AB_hydrolase
IPR004117 7tm6_olfct_rcpt
IPR013818 Lipase/vitellogenin
IPR033906 Lipase_N
IPR000595 cNMP-bd_dom
IPR018488 cNMP-bd_CS
IPR005821 Ion_trans_dom
IPR014710 RmlC-like_jellyroll
IPR018490 cNMP-bd-like
IPR001440 TPR_1
IPR007277 Svp26/Tex261
IPR036867 R3H_dom_sf
IPR011990 TPR-like_helical_dom_sf
IPR013026 TPR-contain_dom
IPR042282 FKBP6/shu
IPR013105 TPR_2
IPR001179 PPIase_FKBP_dom
IPR002048 EF_hand_dom
IPR011992 EF-hand-dom_pair
IPR018247 EF_Hand_1_Ca_BS
IPR000734 TAG_lipase
IPR029058 AB_hydrolase
IPR004117 7tm6_olfct_rcpt
IPR013818 Lipase/vitellogenin
IPR033906 Lipase_N
IPR000595 cNMP-bd_dom
IPR018488 cNMP-bd_CS
IPR005821 Ion_trans_dom
IPR014710 RmlC-like_jellyroll
IPR018490 cNMP-bd-like
IPR001440 TPR_1
IPR007277 Svp26/Tex261
IPR036867 R3H_dom_sf
ProteinModelPortal
H9IWW7
A0A2W1B4N3
A0A0N1I7G7
A0A2A4JPU8
A0A0L7L198
E2A4Y8
+ More
A0A3L8DXT2 A0A026X467 A0A1Y9H221 A0A1X7V8C2 A0A310SIH5 A0A0L0C9J3 T1PHG5 A0A1I8NAG1 F4WY20 A0A0A1XP56 A0A154PEE0 A0A195AYQ9 A0A158NYL1 A0A195DEB8 A0A0J7L1A3 F4X2D5 A0A151WU13 A0A0L7QY84 A0A1B0BNJ8 A0A1A9Y0M9 A0A195FTK4 A0A0K8WCQ6 W8CDP5 A0A0M9A1A3 A0A1A9WIQ6 A0A1I8NY75 A0A2A3EFJ9 A0A034WCL6 A0A182Q1X1 A0A151IEV6 A0A182G8A7 A0A1Q3F193 A0A084VZE6 A0A1A9ZWD6 A0A1B0G7Q4 A0A240PJX4 A0A182R8Q0 A0A182VMP2 E9JDF4 Q16PH6 A0A1S4FTS0 A0A182WF59 A0A1A9V8D6 B0WGW1 U5ENS3 W5JRX6 A0A182JWF1 A0A084VV77 A0A182XCA1 T2MIQ4 A0A182LPI6 A0A182FTD7 E2C0E2 A0A182MGT8 A0A182P9C6 A0A182HNK2 A0A182TN62 A0NFE6 A0A084VU23 A0A2M4BRI7 A0A182T071 A0A146LVU3 A0A182YPF5 B4PA75 A0A194R279 A0A0A9YHD0 A0A0P4W9F9 A0A2L2YF89 A0A1B6F1X8 A0A2B4RN23 A0A023GMQ0 A0A336LDM3 A0A336L5J2 A0A069DYT9
A0A3L8DXT2 A0A026X467 A0A1Y9H221 A0A1X7V8C2 A0A310SIH5 A0A0L0C9J3 T1PHG5 A0A1I8NAG1 F4WY20 A0A0A1XP56 A0A154PEE0 A0A195AYQ9 A0A158NYL1 A0A195DEB8 A0A0J7L1A3 F4X2D5 A0A151WU13 A0A0L7QY84 A0A1B0BNJ8 A0A1A9Y0M9 A0A195FTK4 A0A0K8WCQ6 W8CDP5 A0A0M9A1A3 A0A1A9WIQ6 A0A1I8NY75 A0A2A3EFJ9 A0A034WCL6 A0A182Q1X1 A0A151IEV6 A0A182G8A7 A0A1Q3F193 A0A084VZE6 A0A1A9ZWD6 A0A1B0G7Q4 A0A240PJX4 A0A182R8Q0 A0A182VMP2 E9JDF4 Q16PH6 A0A1S4FTS0 A0A182WF59 A0A1A9V8D6 B0WGW1 U5ENS3 W5JRX6 A0A182JWF1 A0A084VV77 A0A182XCA1 T2MIQ4 A0A182LPI6 A0A182FTD7 E2C0E2 A0A182MGT8 A0A182P9C6 A0A182HNK2 A0A182TN62 A0NFE6 A0A084VU23 A0A2M4BRI7 A0A182T071 A0A146LVU3 A0A182YPF5 B4PA75 A0A194R279 A0A0A9YHD0 A0A0P4W9F9 A0A2L2YF89 A0A1B6F1X8 A0A2B4RN23 A0A023GMQ0 A0A336LDM3 A0A336L5J2 A0A069DYT9
PDB
3B7X
E-value=3.01061e-10,
Score=157
Ontologies
GO
GO:0051879
GO:0051321
GO:0005737
GO:0048477
GO:0031047
GO:0003755
GO:0005509
GO:0016853
GO:0005576
GO:0005549
GO:0006629
GO:0052689
GO:0016021
GO:0004984
GO:0005216
GO:0003676
GO:0070725
GO:0043186
GO:0034587
GO:0005515
GO:0006457
GO:0005525
GO:0005874
GO:0007017
GO:0004559
GO:0006013
GO:0015923
GO:0005634
GO:0006260
Topology
Subcellular location
Cytoplasm
Secreted
Secreted
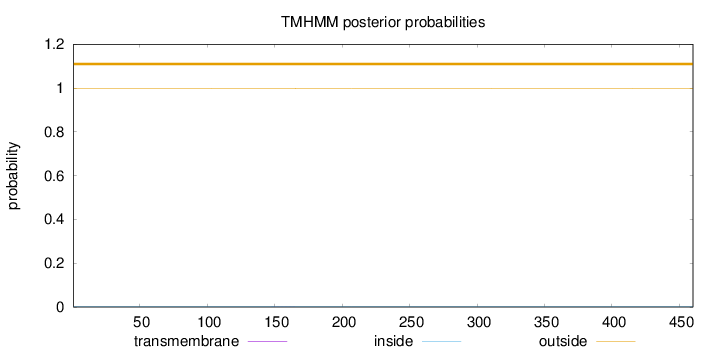
Length:
460
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00345
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00277
outside
1 - 460
Population Genetic Test Statistics
Pi
155.025519
Theta
129.643366
Tajima's D
1.176768
CLR
0.518831
CSRT
0.705014749262537
Interpretation
Uncertain
