Gene
KWMTBOMO06136 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001745
Annotation
PREDICTED:_venom_protease-like_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 3.07
Sequence
CDS
ATGTTGCCTTACGGATTCCAATTACTTTTCCTTGTTTATTTAATAAGAGAAACAGGATGTCAAATAAACGAGGGCGACGTCTGCACGGAGGCCTACACAAACTCAGCCGGGAAATGCACACCGGCGGATACATGTCGATCAGCTAAAGAGGATTTTGTTCAGAACGGCATTAGACCAACGTTTTGCGCCTATACTACCTTCGGAATAGCTCTAGTTTGCTGTCGTGATGGAAGCAGTATACTTCAAACGCCGCCTTCGAGATTAGAAAGTGCTCCTAATGTTTGGGATACGACGGGCAATACAAAAAGGACCAGTGAAAGAAAGTGCGAGGAGTACAGCCGTGGCGTGGTCGAGAAGGTGGACTACCTTCCGCTTCTGCCCGACCCGGATATCCTGTCCATATCGGCGGCGAAATGCGATTACACCGGAATCAAACTGATTGTCGGCGGAGAGAACGCTAATAACGGGGAATTCCCGCATATGGCCGCCATAGGTTGGACTAATTTCGAAGGCTCCTACACCTTCTCCTGCGGGGGCAGCCTCATCAGTCCCCGATTCGTGCTGACGGCCGGACACTGCTCCAGCAACCCCCAGGCCAAGGACCCGGAACCCGTGATCGTGAGGCTGGGGGACCAAAATATAGACCCCACCGTCGACGACGGAGCCAGCCCCATTGATGTACCGATACGGAAGATCAACAAGCACCCGGAATACGCTCCACCGATGGTCTACAACGACATAGCACTCCTGGAGCTCGCTACTGACGTGGAGTTCTCGGCAGCCATACGACCAGCTTGCCTGTGGACGCGGCAGGACTTCGGAGACCACGACAAGGCTCTGGCCACGGGCTGGGGAGTCACTAACACAGAGACGAGAGAAACCGCCAAGGAGCTGCAGAAGGTGTCATTATCGCTACTACAGAATGAATACTGTGATGGTATACTGGAAGCGATTAGAAACAGGAGGTGGCAAGGGTTCGCGGCGACGCAGATGTGCGCTGGCGAGCTGAGGGGAGGCAAGGATACATGTCAGGGCGACTCTGGATCTCCCCTTCAAGTGGCTTCGAAGGATAACCAGTGTATCTTCCACGTTGTTGGAGTGACGTCGTTCGGACGTCGATGCGCCGAGAGCGGTTACCCGGCGATCTACACGAGGGTTGCCAGCTTCATAGACTGGATCGAGAGCGTAGTGTGGCCGGGAGAATAA
Protein
MLPYGFQLLFLVYLIRETGCQINEGDVCTEAYTNSAGKCTPADTCRSAKEDFVQNGIRPTFCAYTTFGIALVCCRDGSSILQTPPSRLESAPNVWDTTGNTKRTSERKCEEYSRGVVEKVDYLPLLPDPDILSISAAKCDYTGIKLIVGGENANNGEFPHMAAIGWTNFEGSYTFSCGGSLISPRFVLTAGHCSSNPQAKDPEPVIVRLGDQNIDPTVDDGASPIDVPIRKINKHPEYAPPMVYNDIALLELATDVEFSAAIRPACLWTRQDFGDHDKALATGWGVTNTETRETAKELQKVSLSLLQNEYCDGILEAIRNRRWQGFAATQMCAGELRGGKDTCQGDSGSPLQVASKDNQCIFHVVGVTSFGRRCAESGYPAIYTRVASFIDWIESVVWPGE
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9IWW2
A0A2A4JQ23
A0A2W1B5H2
G9F9H9
A0A2H1VIS4
A0A212F1E7
+ More
I4DMI4 A0A194Q611 I4DK18 A0A194Q4J1 A0A0N1PHM7 A0A0N0PBX6 Q9XY63 A0A1J1J158 A0A0T6B1X0 A0A182N5I1 A0A1Y9HF60 Q171M7 A0A182LU74 A0A0P6ISZ8 J9HYX9 A0A2J7RDN5 A0A1S4FH91 A0A2J7RDN0 A0A067RX29 A0A182JNP3 A0A182QVG9 A0A2M4DQE4 A0A2M4BQW4 A0A084W6V8 A0A182XWS6 A0A1B0CS99 A0A182L6V0 Q9Y1K5 Q7QAC0 A0A2M4AP36 A0A182X8T1 A0A2M4DQF4 A0A182UYI4 A0A1W4XC70 A0A1L8DQW0 D6W6R5 A0A182U5J8 A0A182VY34 A0A023EQX6 A0A023ESS7 A0A1S4FGZ3 A0A0P6IUV8 A0A023EPR9 I4DLF7 A0A1V1G565 B0W6K2 A0A212EPD4 A0A1Q3FLB4 A0A1B0D6M5 A0A1B6CWC8 W5JGF3 N6UDR4 A0A1B6GJB7 A0A212F1T1 E0V917 W5JJG9 A0A182JI89 A0A2A4JFQ7 Q5MPC5 A0A1S4FI15 A0A182JGW6 A0A3S2NHC8 A0A2H1W7F8 A0A182ID15 A0A182X8T2 Q16ZZ9 U5ERT1 A0A1Q3FNX1 Q16PM8 A0A182VFE5 A0A023EPF2 A0A1S4FU89 A0A194R9C5 A0A1Y9HED1 X1WIF2 A0A0P6IV39 A0A182H431 A0A1S4FTN1 A0A194RAB5 A0A0M8ZW49 A0A1Y1MWZ7 A0A1I8NBH8 B0W1A9 Q170A0 A0A023EQW3 A0A182GIP9 A0A182JNP4
I4DMI4 A0A194Q611 I4DK18 A0A194Q4J1 A0A0N1PHM7 A0A0N0PBX6 Q9XY63 A0A1J1J158 A0A0T6B1X0 A0A182N5I1 A0A1Y9HF60 Q171M7 A0A182LU74 A0A0P6ISZ8 J9HYX9 A0A2J7RDN5 A0A1S4FH91 A0A2J7RDN0 A0A067RX29 A0A182JNP3 A0A182QVG9 A0A2M4DQE4 A0A2M4BQW4 A0A084W6V8 A0A182XWS6 A0A1B0CS99 A0A182L6V0 Q9Y1K5 Q7QAC0 A0A2M4AP36 A0A182X8T1 A0A2M4DQF4 A0A182UYI4 A0A1W4XC70 A0A1L8DQW0 D6W6R5 A0A182U5J8 A0A182VY34 A0A023EQX6 A0A023ESS7 A0A1S4FGZ3 A0A0P6IUV8 A0A023EPR9 I4DLF7 A0A1V1G565 B0W6K2 A0A212EPD4 A0A1Q3FLB4 A0A1B0D6M5 A0A1B6CWC8 W5JGF3 N6UDR4 A0A1B6GJB7 A0A212F1T1 E0V917 W5JJG9 A0A182JI89 A0A2A4JFQ7 Q5MPC5 A0A1S4FI15 A0A182JGW6 A0A3S2NHC8 A0A2H1W7F8 A0A182ID15 A0A182X8T2 Q16ZZ9 U5ERT1 A0A1Q3FNX1 Q16PM8 A0A182VFE5 A0A023EPF2 A0A1S4FU89 A0A194R9C5 A0A1Y9HED1 X1WIF2 A0A0P6IV39 A0A182H431 A0A1S4FTN1 A0A194RAB5 A0A0M8ZW49 A0A1Y1MWZ7 A0A1I8NBH8 B0W1A9 Q170A0 A0A023EQW3 A0A182GIP9 A0A182JNP4
Pubmed
EMBL
BABH01021026
NWSH01000865
PCG73808.1
KZ150313
PZC71449.1
JN033724
+ More
AEW46877.1 ODYU01002797 SOQ40740.1 AGBW02010907 OWR47533.1 AK402502 BAM19124.1 KQ459463 KPJ00445.1 AK401636 BAM18258.1 KPJ00462.1 KQ458725 KPJ05344.1 KQ460736 KPJ12593.1 AF053921 AAD21841.1 CVRI01000064 CRL05174.1 LJIG01016282 KRT81069.1 CH477450 EAT40694.2 AXCM01003236 GDUN01001005 JAN94914.1 EJY57695.1 NEVH01005287 PNF38945.1 PNF38947.1 KK852411 KDR24454.1 AXCN02001168 GGFL01015594 MBW79772.1 GGFJ01006253 MBW55394.1 ATLV01020986 KE525311 KFB45952.1 AJWK01025808 AJWK01025809 AF117750 AAD38336.1 AAAB01008898 EAA09190.2 GGFK01009233 MBW42554.1 GGFL01015607 MBW79785.1 GFDF01005349 JAV08735.1 KQ971307 EFA11559.1 GAPW01002075 JAC11523.1 GAPW01001752 JAC11846.1 GDUN01001000 JAN94919.1 GAPW01002076 JAC11522.1 AK402125 BAM18747.1 FX985323 BAX07336.1 DS231848 EDS36676.1 AGBW02013495 OWR43336.1 GFDL01006707 JAV28338.1 AJVK01012092 GEDC01019518 GEDC01019298 JAS17780.1 JAS18000.1 ADMH02001254 ETN63437.1 APGK01039086 APGK01039087 APGK01039088 KB740967 ENN76802.1 GECZ01007233 GECZ01005940 JAS62536.1 JAS63829.1 AGBW02010816 OWR47692.1 DS234988 EEB09873.1 ETN63438.1 NWSH01001694 PCG70404.1 AY672785 AAV91007.1 RSAL01000098 RVE47611.1 ODYU01006800 SOQ48987.1 APCN01004968 CH477477 EAT40291.1 GANO01003579 JAB56292.1 GFDL01005857 JAV29188.1 CH477777 EAT36311.1 GAPW01002718 JAC10880.1 KQ460473 KPJ14443.1 ABLF02031950 ABLF02031955 GDUN01000999 JAN94920.1 JXUM01108692 KQ565255 KXJ71168.1 KPJ14444.1 KQ435821 KOX72340.1 GEZM01021493 JAV88975.1 DS231821 EDS44979.1 EAT40292.1 GAPW01002090 JAC11508.1 JXUM01012146 KQ560346 KXJ82932.1
AEW46877.1 ODYU01002797 SOQ40740.1 AGBW02010907 OWR47533.1 AK402502 BAM19124.1 KQ459463 KPJ00445.1 AK401636 BAM18258.1 KPJ00462.1 KQ458725 KPJ05344.1 KQ460736 KPJ12593.1 AF053921 AAD21841.1 CVRI01000064 CRL05174.1 LJIG01016282 KRT81069.1 CH477450 EAT40694.2 AXCM01003236 GDUN01001005 JAN94914.1 EJY57695.1 NEVH01005287 PNF38945.1 PNF38947.1 KK852411 KDR24454.1 AXCN02001168 GGFL01015594 MBW79772.1 GGFJ01006253 MBW55394.1 ATLV01020986 KE525311 KFB45952.1 AJWK01025808 AJWK01025809 AF117750 AAD38336.1 AAAB01008898 EAA09190.2 GGFK01009233 MBW42554.1 GGFL01015607 MBW79785.1 GFDF01005349 JAV08735.1 KQ971307 EFA11559.1 GAPW01002075 JAC11523.1 GAPW01001752 JAC11846.1 GDUN01001000 JAN94919.1 GAPW01002076 JAC11522.1 AK402125 BAM18747.1 FX985323 BAX07336.1 DS231848 EDS36676.1 AGBW02013495 OWR43336.1 GFDL01006707 JAV28338.1 AJVK01012092 GEDC01019518 GEDC01019298 JAS17780.1 JAS18000.1 ADMH02001254 ETN63437.1 APGK01039086 APGK01039087 APGK01039088 KB740967 ENN76802.1 GECZ01007233 GECZ01005940 JAS62536.1 JAS63829.1 AGBW02010816 OWR47692.1 DS234988 EEB09873.1 ETN63438.1 NWSH01001694 PCG70404.1 AY672785 AAV91007.1 RSAL01000098 RVE47611.1 ODYU01006800 SOQ48987.1 APCN01004968 CH477477 EAT40291.1 GANO01003579 JAB56292.1 GFDL01005857 JAV29188.1 CH477777 EAT36311.1 GAPW01002718 JAC10880.1 KQ460473 KPJ14443.1 ABLF02031950 ABLF02031955 GDUN01000999 JAN94920.1 JXUM01108692 KQ565255 KXJ71168.1 KPJ14444.1 KQ435821 KOX72340.1 GEZM01021493 JAV88975.1 DS231821 EDS44979.1 EAT40292.1 GAPW01002090 JAC11508.1 JXUM01012146 KQ560346 KXJ82932.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000183832
+ More
UP000075884 UP000075900 UP000008820 UP000075883 UP000235965 UP000027135 UP000075881 UP000075886 UP000030765 UP000076408 UP000092461 UP000075882 UP000007062 UP000076407 UP000075903 UP000192223 UP000007266 UP000075902 UP000075920 UP000002320 UP000092462 UP000000673 UP000019118 UP000009046 UP000075880 UP000283053 UP000075840 UP000007819 UP000069940 UP000249989 UP000053105 UP000095301
UP000075884 UP000075900 UP000008820 UP000075883 UP000235965 UP000027135 UP000075881 UP000075886 UP000030765 UP000076408 UP000092461 UP000075882 UP000007062 UP000076407 UP000075903 UP000192223 UP000007266 UP000075902 UP000075920 UP000002320 UP000092462 UP000000673 UP000019118 UP000009046 UP000075880 UP000283053 UP000075840 UP000007819 UP000069940 UP000249989 UP000053105 UP000095301
PRIDE
Pfam
Interpro
IPR018114
TRYPSIN_HIS
+ More
IPR001254 Trypsin_dom
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR033116 TRYPSIN_SER
IPR036236 Znf_C2H2_sf
IPR009668 RNA_pol-assoc_fac_A49-like
IPR013087 Znf_C2H2_type
IPR022700 CLIP
IPR005304 Rbsml_bgen_MeTrfase_EMG1/NEP1
IPR029028 Alpha/beta_knot_MTases
IPR029026 tRNA_m1G_MTases_N
IPR008598 Di19_Zn_binding_dom
IPR001254 Trypsin_dom
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR033116 TRYPSIN_SER
IPR036236 Znf_C2H2_sf
IPR009668 RNA_pol-assoc_fac_A49-like
IPR013087 Znf_C2H2_type
IPR022700 CLIP
IPR005304 Rbsml_bgen_MeTrfase_EMG1/NEP1
IPR029028 Alpha/beta_knot_MTases
IPR029026 tRNA_m1G_MTases_N
IPR008598 Di19_Zn_binding_dom
Gene 3D
CDD
ProteinModelPortal
H9IWW2
A0A2A4JQ23
A0A2W1B5H2
G9F9H9
A0A2H1VIS4
A0A212F1E7
+ More
I4DMI4 A0A194Q611 I4DK18 A0A194Q4J1 A0A0N1PHM7 A0A0N0PBX6 Q9XY63 A0A1J1J158 A0A0T6B1X0 A0A182N5I1 A0A1Y9HF60 Q171M7 A0A182LU74 A0A0P6ISZ8 J9HYX9 A0A2J7RDN5 A0A1S4FH91 A0A2J7RDN0 A0A067RX29 A0A182JNP3 A0A182QVG9 A0A2M4DQE4 A0A2M4BQW4 A0A084W6V8 A0A182XWS6 A0A1B0CS99 A0A182L6V0 Q9Y1K5 Q7QAC0 A0A2M4AP36 A0A182X8T1 A0A2M4DQF4 A0A182UYI4 A0A1W4XC70 A0A1L8DQW0 D6W6R5 A0A182U5J8 A0A182VY34 A0A023EQX6 A0A023ESS7 A0A1S4FGZ3 A0A0P6IUV8 A0A023EPR9 I4DLF7 A0A1V1G565 B0W6K2 A0A212EPD4 A0A1Q3FLB4 A0A1B0D6M5 A0A1B6CWC8 W5JGF3 N6UDR4 A0A1B6GJB7 A0A212F1T1 E0V917 W5JJG9 A0A182JI89 A0A2A4JFQ7 Q5MPC5 A0A1S4FI15 A0A182JGW6 A0A3S2NHC8 A0A2H1W7F8 A0A182ID15 A0A182X8T2 Q16ZZ9 U5ERT1 A0A1Q3FNX1 Q16PM8 A0A182VFE5 A0A023EPF2 A0A1S4FU89 A0A194R9C5 A0A1Y9HED1 X1WIF2 A0A0P6IV39 A0A182H431 A0A1S4FTN1 A0A194RAB5 A0A0M8ZW49 A0A1Y1MWZ7 A0A1I8NBH8 B0W1A9 Q170A0 A0A023EQW3 A0A182GIP9 A0A182JNP4
I4DMI4 A0A194Q611 I4DK18 A0A194Q4J1 A0A0N1PHM7 A0A0N0PBX6 Q9XY63 A0A1J1J158 A0A0T6B1X0 A0A182N5I1 A0A1Y9HF60 Q171M7 A0A182LU74 A0A0P6ISZ8 J9HYX9 A0A2J7RDN5 A0A1S4FH91 A0A2J7RDN0 A0A067RX29 A0A182JNP3 A0A182QVG9 A0A2M4DQE4 A0A2M4BQW4 A0A084W6V8 A0A182XWS6 A0A1B0CS99 A0A182L6V0 Q9Y1K5 Q7QAC0 A0A2M4AP36 A0A182X8T1 A0A2M4DQF4 A0A182UYI4 A0A1W4XC70 A0A1L8DQW0 D6W6R5 A0A182U5J8 A0A182VY34 A0A023EQX6 A0A023ESS7 A0A1S4FGZ3 A0A0P6IUV8 A0A023EPR9 I4DLF7 A0A1V1G565 B0W6K2 A0A212EPD4 A0A1Q3FLB4 A0A1B0D6M5 A0A1B6CWC8 W5JGF3 N6UDR4 A0A1B6GJB7 A0A212F1T1 E0V917 W5JJG9 A0A182JI89 A0A2A4JFQ7 Q5MPC5 A0A1S4FI15 A0A182JGW6 A0A3S2NHC8 A0A2H1W7F8 A0A182ID15 A0A182X8T2 Q16ZZ9 U5ERT1 A0A1Q3FNX1 Q16PM8 A0A182VFE5 A0A023EPF2 A0A1S4FU89 A0A194R9C5 A0A1Y9HED1 X1WIF2 A0A0P6IV39 A0A182H431 A0A1S4FTN1 A0A194RAB5 A0A0M8ZW49 A0A1Y1MWZ7 A0A1I8NBH8 B0W1A9 Q170A0 A0A023EQW3 A0A182GIP9 A0A182JNP4
PDB
2OLG
E-value=3.88442e-31,
Score=337
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.981043
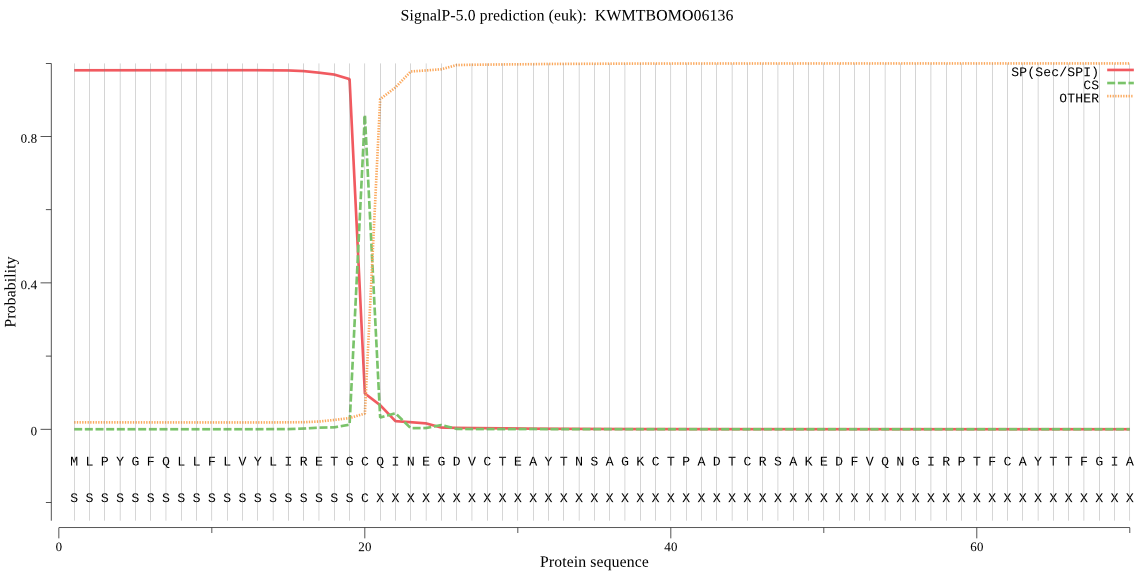
Length:
401
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00519
Exp number, first 60 AAs:
0.00098
Total prob of N-in:
0.00040
outside
1 - 401
Population Genetic Test Statistics
Pi
180.150652
Theta
146.399473
Tajima's D
0.593211
CLR
0.431394
CSRT
0.535773211339433
Interpretation
Uncertain
