Gene
KWMTBOMO06133
Pre Gene Modal
BGIBMGA001743
Annotation
PREDICTED:_zinc_finger_protein_271-like_isoform_X2_[Bombyx_mori]
Full name
Zinc finger protein 22
Alternative Name
Zinc finger protein KOX15
Zinc finger protein Krox-26
Zinc finger protein Krox-26
Transcription factor
Location in the cell
Nuclear Reliability : 2.311
Sequence
CDS
ATGTGGGTCCACACCGGCGAGAAACGCTTCAAATGCGACCGCTGCCCGAAGAGCTTCACCCAGAAGACGAACCTGGTGTTCCACCTGCGCGTGCACAGCGCCACCCGACCCTCGTACGAGTGCCCCCTGTGCGGGAAACATTTCGCCTTCTATAACAACAGGAGGCGTCATATGTTCATCCACACTGGCCTGAAGCCGTACAAATGCGACACGTGCCTCAAGTGCTTCACAACGTCCGGCGAGCTGCGCGCGCACGTCGAGCACGTGCACATGAAGAAGCCGTGGCCGAAGCGCGCGCGTACCAACAGACAGAGCGACGCGCTCGCCGACTAG
Protein
MWVHTGEKRFKCDRCPKSFTQKTNLVFHLRVHSATRPSYECPLCGKHFAFYNNRRRHMFIHTGLKPYKCDTCLKCFTTSGELRAHVEHVHMKKPWPKRARTNRQSDALAD
Summary
Description
Binds DNA through the consensus sequence 5'-CAATG-3'. May be involved in transcriptional regulation and may play a role in tooth formation (By similarity).
Similarity
Belongs to the krueppel C2H2-type zinc-finger protein family.
Keywords
Acetylation
Complete proteome
DNA-binding
Metal-binding
Nucleus
Phosphoprotein
Reference proteome
Repeat
Transcription
Transcription regulation
Zinc
Zinc-finger
Polymorphism
Feature
chain Zinc finger protein 22
sequence variant In dbSNP:rs3740093.
sequence variant In dbSNP:rs3740093.
Uniprot
H9IWW0
A0A2A4KA39
A0A3S2TJB5
A0A2H1W3X2
A0A194Q527
A0A194QVL0
+ More
A0A1E1WHH4 A0A2A4K8P9 A0A2W1BJN6 A0A194QRI4 A0A194Q632 A0A2A4JSJ1 H9IWW1 A0A2A4JTI5 A0A212ET42 A0A212F5H6 A0A2H1VJD2 A0A0L7LGN8 A0A0L7LH80 A0A2A4K955 A0A2H1VTH9 S4PE06 S4PEJ9 A0A2H1W0Z4 A0A2D4GJC2 A7SWT3 A0A2D4PD48 Q8VEC1 A0A3B2W7H5 A7S0X6 A7T2W2 E9Q5G8 S4P1V0 A0A091DUI9 A0A3Q0ECV2 V4A5U0 A0A2A4JTE3 A0A061IJP8 A0A3Q3IL18 A0A0L8HUG3 A0A0L8HUD4 A0A3N0ZA52 A0A0P6JD94 A0A0L8H6P6 A0A0L8IAJ3 A0A3Q7MCV9 A0A3B4BJ45 A0A1W4XVQ6 A0A1W4XV68 A0A1W4XV63 A7T1L9 A0A3P8NTM5 A7TAT8 A0A1W4Y5B1 A0A1W4Y4W3 A0A1W4Y5G4 A0A3P9DGF5 A0A3B4F2M7 A0A2B4REX0 K1PPH7 A0A0K8W733 K9KBQ3 A0A2W1BLU1 A0A0L8HTA0 A0A0K8U4S7 A0A3B5A840 A0A1U7SUI7 G3IEA2 A0A3Q1ECG0 A0A3B5LI64 A0A3N0YZ26 A0A0R4IXX9 A0A0L8HV31 B7ZUX7 V4AAN6 A0A3Q0DM75 A8WFV9 A7T4M0 A0A2R8ZMM4 A0A3Q2Q910 A0A146VLS6 A0A0L8I532 U3AYV7 A0A2K5RJT2 A0A2I3HJF5 A0A146ZSC1 F6Y801 A0A3Q1FXM1 A0A0L8FS27 A0A2I2YAN2 A0A2J8XT63 H2Q1U7 A0A024R7T4 Q5R4X5 P17026 A0A3Q1H518 A0A0L8H224 A0A3P9CKJ5 A0A0L8GTC8
A0A1E1WHH4 A0A2A4K8P9 A0A2W1BJN6 A0A194QRI4 A0A194Q632 A0A2A4JSJ1 H9IWW1 A0A2A4JTI5 A0A212ET42 A0A212F5H6 A0A2H1VJD2 A0A0L7LGN8 A0A0L7LH80 A0A2A4K955 A0A2H1VTH9 S4PE06 S4PEJ9 A0A2H1W0Z4 A0A2D4GJC2 A7SWT3 A0A2D4PD48 Q8VEC1 A0A3B2W7H5 A7S0X6 A7T2W2 E9Q5G8 S4P1V0 A0A091DUI9 A0A3Q0ECV2 V4A5U0 A0A2A4JTE3 A0A061IJP8 A0A3Q3IL18 A0A0L8HUG3 A0A0L8HUD4 A0A3N0ZA52 A0A0P6JD94 A0A0L8H6P6 A0A0L8IAJ3 A0A3Q7MCV9 A0A3B4BJ45 A0A1W4XVQ6 A0A1W4XV68 A0A1W4XV63 A7T1L9 A0A3P8NTM5 A7TAT8 A0A1W4Y5B1 A0A1W4Y4W3 A0A1W4Y5G4 A0A3P9DGF5 A0A3B4F2M7 A0A2B4REX0 K1PPH7 A0A0K8W733 K9KBQ3 A0A2W1BLU1 A0A0L8HTA0 A0A0K8U4S7 A0A3B5A840 A0A1U7SUI7 G3IEA2 A0A3Q1ECG0 A0A3B5LI64 A0A3N0YZ26 A0A0R4IXX9 A0A0L8HV31 B7ZUX7 V4AAN6 A0A3Q0DM75 A8WFV9 A7T4M0 A0A2R8ZMM4 A0A3Q2Q910 A0A146VLS6 A0A0L8I532 U3AYV7 A0A2K5RJT2 A0A2I3HJF5 A0A146ZSC1 F6Y801 A0A3Q1FXM1 A0A0L8FS27 A0A2I2YAN2 A0A2J8XT63 H2Q1U7 A0A024R7T4 Q5R4X5 P17026 A0A3Q1H518 A0A0L8H224 A0A3P9CKJ5 A0A0L8GTC8
Pubmed
EMBL
BABH01021017
BABH01021018
BABH01021019
NWSH01000027
PCG80590.1
RSAL01000098
+ More
RVE47648.1 ODYU01006174 SOQ47799.1 KQ459463 KPJ00464.1 KQ461181 KPJ07591.1 GDQN01004610 JAT86444.1 PCG80587.1 KZ150245 PZC71883.1 KPJ07590.1 KPJ00465.1 NWSH01000652 PCG75007.1 BABH01021020 BABH01021021 BABH01021022 BABH01021023 PCG75008.1 AGBW02012649 OWR44658.1 AGBW02010195 OWR48987.1 ODYU01002887 SOQ40937.1 JTDY01001260 KOB74361.1 JTDY01001125 KOB74797.1 PCG80589.1 ODYU01004320 SOQ44066.1 GAIX01001789 JAA90771.1 GAIX01001539 JAA91021.1 ODYU01005670 SOQ46765.1 IACJ01133402 LAA59819.1 DS469867 EDO31838.1 IACN01068181 LAB55950.1 BC019219 AAH19219.1 CT009755 DS469562 EDO42660.1 DS470341 EDO29702.1 GAIX01012305 JAA80255.1 KN122033 KFO33955.1 KB202444 ESO90340.1 NWSH01000707 PCG74670.1 KE667083 ERE87554.1 KQ417280 KOF92851.1 KOF92821.1 RJVU01001269 ROL55261.1 GEBF01001268 JAO02365.1 KQ419049 KOF84759.1 KQ416225 KOF98035.1 DS470123 EDO30148.1 DS474437 EDO26879.1 LSMT01000709 PFX14928.1 JH816467 EKC18365.1 GDHF01005373 JAI46941.1 JL621397 AEP99455.1 KZ150166 PZC72653.1 KQ417336 KOF92441.1 GDHF01031004 JAI21310.1 JH002160 EGW13504.1 RJVU01019022 ROL51262.1 BX004964 BX248582 KQ417236 KOF93064.1 BC171385 AAI71385.1 ESO90341.1 BC154476 AAI54477.1 DS470906 EDO29094.1 AJFE02056893 GCES01068426 JAR17897.1 KQ416560 KOF96464.1 GAMT01006389 GAMT01006388 GAMS01007182 GAMS01007181 GAMR01009212 GAMR01009211 GAMQ01003414 GAMQ01003413 GAMP01004019 GAMP01004018 JAB05473.1 JAB15954.1 JAB24720.1 JAB38437.1 JAB48736.1 ADFV01064219 GCES01017225 JAR69098.1 KQ427054 KOF67463.1 CABD030071137 NDHI03003318 PNJ85226.1 AACZ04049673 GABC01010476 GABF01005519 GABD01007771 GABE01003637 GABE01003636 NBAG03000468 JAA00862.1 JAA16626.1 JAA25329.1 JAA41102.1 PNI20895.1 CH471160 EAW86633.1 CR861114 S50223 AY137767 BT009806 AK291508 AL353801 BC010642 BC041139 BC053687 M77172 X52346 KQ419523 KOF83316.1 KQ420458 KOF80213.1
RVE47648.1 ODYU01006174 SOQ47799.1 KQ459463 KPJ00464.1 KQ461181 KPJ07591.1 GDQN01004610 JAT86444.1 PCG80587.1 KZ150245 PZC71883.1 KPJ07590.1 KPJ00465.1 NWSH01000652 PCG75007.1 BABH01021020 BABH01021021 BABH01021022 BABH01021023 PCG75008.1 AGBW02012649 OWR44658.1 AGBW02010195 OWR48987.1 ODYU01002887 SOQ40937.1 JTDY01001260 KOB74361.1 JTDY01001125 KOB74797.1 PCG80589.1 ODYU01004320 SOQ44066.1 GAIX01001789 JAA90771.1 GAIX01001539 JAA91021.1 ODYU01005670 SOQ46765.1 IACJ01133402 LAA59819.1 DS469867 EDO31838.1 IACN01068181 LAB55950.1 BC019219 AAH19219.1 CT009755 DS469562 EDO42660.1 DS470341 EDO29702.1 GAIX01012305 JAA80255.1 KN122033 KFO33955.1 KB202444 ESO90340.1 NWSH01000707 PCG74670.1 KE667083 ERE87554.1 KQ417280 KOF92851.1 KOF92821.1 RJVU01001269 ROL55261.1 GEBF01001268 JAO02365.1 KQ419049 KOF84759.1 KQ416225 KOF98035.1 DS470123 EDO30148.1 DS474437 EDO26879.1 LSMT01000709 PFX14928.1 JH816467 EKC18365.1 GDHF01005373 JAI46941.1 JL621397 AEP99455.1 KZ150166 PZC72653.1 KQ417336 KOF92441.1 GDHF01031004 JAI21310.1 JH002160 EGW13504.1 RJVU01019022 ROL51262.1 BX004964 BX248582 KQ417236 KOF93064.1 BC171385 AAI71385.1 ESO90341.1 BC154476 AAI54477.1 DS470906 EDO29094.1 AJFE02056893 GCES01068426 JAR17897.1 KQ416560 KOF96464.1 GAMT01006389 GAMT01006388 GAMS01007182 GAMS01007181 GAMR01009212 GAMR01009211 GAMQ01003414 GAMQ01003413 GAMP01004019 GAMP01004018 JAB05473.1 JAB15954.1 JAB24720.1 JAB38437.1 JAB48736.1 ADFV01064219 GCES01017225 JAR69098.1 KQ427054 KOF67463.1 CABD030071137 NDHI03003318 PNJ85226.1 AACZ04049673 GABC01010476 GABF01005519 GABD01007771 GABE01003637 GABE01003636 NBAG03000468 JAA00862.1 JAA16626.1 JAA25329.1 JAA41102.1 PNI20895.1 CH471160 EAW86633.1 CR861114 S50223 AY137767 BT009806 AK291508 AL353801 BC010642 BC041139 BC053687 M77172 X52346 KQ419523 KOF83316.1 KQ420458 KOF80213.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000037510 UP000001593 UP000000589 UP000028990 UP000189704 UP000030746 UP000030759 UP000261600 UP000053454 UP000286641 UP000261520 UP000192224 UP000265100 UP000265160 UP000261460 UP000225706 UP000005408 UP000261400 UP000001075 UP000257200 UP000261380 UP000000437 UP000240080 UP000265000 UP000008225 UP000233040 UP000001073 UP000002281 UP000001519 UP000002277 UP000001595 UP000005640 UP000265040
UP000037510 UP000001593 UP000000589 UP000028990 UP000189704 UP000030746 UP000030759 UP000261600 UP000053454 UP000286641 UP000261520 UP000192224 UP000265100 UP000265160 UP000261460 UP000225706 UP000005408 UP000261400 UP000001075 UP000257200 UP000261380 UP000000437 UP000240080 UP000265000 UP000008225 UP000233040 UP000001073 UP000002281 UP000001519 UP000002277 UP000001595 UP000005640 UP000265040
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9IWW0
A0A2A4KA39
A0A3S2TJB5
A0A2H1W3X2
A0A194Q527
A0A194QVL0
+ More
A0A1E1WHH4 A0A2A4K8P9 A0A2W1BJN6 A0A194QRI4 A0A194Q632 A0A2A4JSJ1 H9IWW1 A0A2A4JTI5 A0A212ET42 A0A212F5H6 A0A2H1VJD2 A0A0L7LGN8 A0A0L7LH80 A0A2A4K955 A0A2H1VTH9 S4PE06 S4PEJ9 A0A2H1W0Z4 A0A2D4GJC2 A7SWT3 A0A2D4PD48 Q8VEC1 A0A3B2W7H5 A7S0X6 A7T2W2 E9Q5G8 S4P1V0 A0A091DUI9 A0A3Q0ECV2 V4A5U0 A0A2A4JTE3 A0A061IJP8 A0A3Q3IL18 A0A0L8HUG3 A0A0L8HUD4 A0A3N0ZA52 A0A0P6JD94 A0A0L8H6P6 A0A0L8IAJ3 A0A3Q7MCV9 A0A3B4BJ45 A0A1W4XVQ6 A0A1W4XV68 A0A1W4XV63 A7T1L9 A0A3P8NTM5 A7TAT8 A0A1W4Y5B1 A0A1W4Y4W3 A0A1W4Y5G4 A0A3P9DGF5 A0A3B4F2M7 A0A2B4REX0 K1PPH7 A0A0K8W733 K9KBQ3 A0A2W1BLU1 A0A0L8HTA0 A0A0K8U4S7 A0A3B5A840 A0A1U7SUI7 G3IEA2 A0A3Q1ECG0 A0A3B5LI64 A0A3N0YZ26 A0A0R4IXX9 A0A0L8HV31 B7ZUX7 V4AAN6 A0A3Q0DM75 A8WFV9 A7T4M0 A0A2R8ZMM4 A0A3Q2Q910 A0A146VLS6 A0A0L8I532 U3AYV7 A0A2K5RJT2 A0A2I3HJF5 A0A146ZSC1 F6Y801 A0A3Q1FXM1 A0A0L8FS27 A0A2I2YAN2 A0A2J8XT63 H2Q1U7 A0A024R7T4 Q5R4X5 P17026 A0A3Q1H518 A0A0L8H224 A0A3P9CKJ5 A0A0L8GTC8
A0A1E1WHH4 A0A2A4K8P9 A0A2W1BJN6 A0A194QRI4 A0A194Q632 A0A2A4JSJ1 H9IWW1 A0A2A4JTI5 A0A212ET42 A0A212F5H6 A0A2H1VJD2 A0A0L7LGN8 A0A0L7LH80 A0A2A4K955 A0A2H1VTH9 S4PE06 S4PEJ9 A0A2H1W0Z4 A0A2D4GJC2 A7SWT3 A0A2D4PD48 Q8VEC1 A0A3B2W7H5 A7S0X6 A7T2W2 E9Q5G8 S4P1V0 A0A091DUI9 A0A3Q0ECV2 V4A5U0 A0A2A4JTE3 A0A061IJP8 A0A3Q3IL18 A0A0L8HUG3 A0A0L8HUD4 A0A3N0ZA52 A0A0P6JD94 A0A0L8H6P6 A0A0L8IAJ3 A0A3Q7MCV9 A0A3B4BJ45 A0A1W4XVQ6 A0A1W4XV68 A0A1W4XV63 A7T1L9 A0A3P8NTM5 A7TAT8 A0A1W4Y5B1 A0A1W4Y4W3 A0A1W4Y5G4 A0A3P9DGF5 A0A3B4F2M7 A0A2B4REX0 K1PPH7 A0A0K8W733 K9KBQ3 A0A2W1BLU1 A0A0L8HTA0 A0A0K8U4S7 A0A3B5A840 A0A1U7SUI7 G3IEA2 A0A3Q1ECG0 A0A3B5LI64 A0A3N0YZ26 A0A0R4IXX9 A0A0L8HV31 B7ZUX7 V4AAN6 A0A3Q0DM75 A8WFV9 A7T4M0 A0A2R8ZMM4 A0A3Q2Q910 A0A146VLS6 A0A0L8I532 U3AYV7 A0A2K5RJT2 A0A2I3HJF5 A0A146ZSC1 F6Y801 A0A3Q1FXM1 A0A0L8FS27 A0A2I2YAN2 A0A2J8XT63 H2Q1U7 A0A024R7T4 Q5R4X5 P17026 A0A3Q1H518 A0A0L8H224 A0A3P9CKJ5 A0A0L8GTC8
PDB
2I13
E-value=1.32755e-13,
Score=178
Ontologies
GO
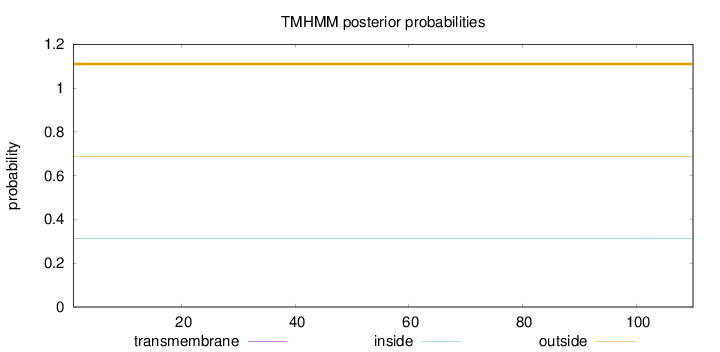
Topology
Subcellular location
Nucleus
Length:
110
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0008
Exp number, first 60 AAs:
0.0008
Total prob of N-in:
0.31215
outside
1 - 110
Population Genetic Test Statistics
Pi
142.247698
Theta
133.173166
Tajima's D
0.207458
CLR
0.626094
CSRT
0.425828708564572
Interpretation
Uncertain
