Gene
KWMTBOMO06114
Pre Gene Modal
BGIBMGA001691
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC101737330_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.735
Sequence
CDS
ATGTCTCCGACGGAGCAGGAGGAGGATTCCCTGCTGACGCTGTCTTCGGTGACGGCGAGGCCGCTGCTGGCCGCGTCCAGGAAGTTGAGACCTCCAGGGTCGAATGCCGAGAGCATCGTCCCTCCCGACGGCGGCTACGGCTGGGTGGTGGTATTCGGCGCGTTCATGGTTCAGTTTTGGGTGGCTGGTCTCTTCAAGTCCTACGGGGTTCTGTACGTGGAGATAATGGAGACGTTTCCGGATTCGTCAGCTTCGGTCGCTTCGTGGATTCCGGCCGCTCTGTCGACGTTGTGCTTGGCTTTAGCACCACTGTCGTCGGCTCTCTGCGAGAAGTACTCCTGCAGGCTGGTCGTGTTCATCGGTGGGCTGTTCTGCGCGAGTGGACTCGCTCTCTCGTTCTTCAGCACGGGACTGCTACATCTGCTGCTCAGTTTTGGGGTCATGACCGGTATCGGCGGAGGACTGTCCACAACGCCAGGCATCGTCATTGTGTCGCAGTACTTCGACAAGCACAGGGCCCTGGCTAATGGTATTTGCGTGAGCGGGACGGCTGCCGGCAGCTTCGTCTTCCCGATGTTGATCGAGAAACTGGTGGATGTATATGGACTGCACGGAACCGTTCTTCTGTTAGGTGGGGGGATGCTGCACGTGTGCGTGGCGGCGACCTTATACCGTCCCCCGCCGGTGGCAGGCTGCGGGGAGCGGACCCCAGCGACCACCACCACGACCGAACACTCCACTCTACTCAACGACATCAACGATGGTGGCAAAATCGAGCCGCTCAAGGACAACTGCCTCCACAGCTTGTTTCACCACAACCCCGAACTGGCCCTGAACCAGAAGAACGTTTTGCTGCAAGAAGAAACGAAAAGAATGAACCTAATGACAGAGATAATGCATCCGAACTTAATACAAGTAGTGCGACAGTCCCAGCTGTCTGACTCTGAAGATTCCGAAGGGATGGACGTCCTCGGAGTGGTCGGTCTACCGCGCGAGGTAGTGAGACTACGCCTCCGGCCGATCAGATCCTCGTCGATCCTTCATTCAGTTGAGGACCTGTCTACAGACAGCACTTGCGTCTACAAGGCTTCGAGAAGGAAATTGAGTCGCTCTCTGTATATTCGTCCGCCCGTGACCAGCCGCTTGACACAGAGTAAGCTCGCCGACCAGGTCATCAGGAAAGAAGTTGAAATTAAGTCCGAAGAACCCGAGAAGAAACCGAGCAAGGGTTGCGTTGAAAAGATATCCAGATACTTGGACGTTTCGCTGTTGAAGTCGTGGCGATTCGGTTTGCTTTGTACGTCTGTGGCGCTGATGTCCTGTGGCTCCCCTTACGCGCTGTACTACTTACCGGCGTACACGGTCGCCATAGGGCACACGAAGGCAGCCGCCGGTCGGTTGGTGGCCATCAGCGCTGTACTGGACCTGTGCGGGAGGCTGGGGCTGGGCTGGCTGTGCGATCTACACTTGTTCTGTAGAAGGAAAGCCTATATCATCAGTATCTTCGTTGCTGGAGCGGCTGTGCTGACGCTGCCGAGTCTAACTTCCTGGTGGTCCCTCGCTATAGTCTCAGGAGCGTACGGCCTCTGCCTCGGCTGCTGGTTCCTGCTGGTTCCGGTGCTGCTGGCGGACGCCTTCGGCACCGCGCGCATTGCGTCCTCGTACGGGCTCGTCAGAATGTTCCAGAGCGTAGCTGCCGTCTCTGTACCACCAACTGCAGGCTATATAAGAGACTTGACCGGAGGTTTCTCCTGGTGCTTCTACGGGATGGGATCCTGCATGGTCCTGGGCACGATCCCGGTCATAGCGTTCCACGTCAGCACCAAGAACGACGACAATGGCTCCACGGTGGACTGA
Protein
MSPTEQEEDSLLTLSSVTARPLLAASRKLRPPGSNAESIVPPDGGYGWVVVFGAFMVQFWVAGLFKSYGVLYVEIMETFPDSSASVASWIPAALSTLCLALAPLSSALCEKYSCRLVVFIGGLFCASGLALSFFSTGLLHLLLSFGVMTGIGGGLSTTPGIVIVSQYFDKHRALANGICVSGTAAGSFVFPMLIEKLVDVYGLHGTVLLLGGGMLHVCVAATLYRPPPVAGCGERTPATTTTTEHSTLLNDINDGGKIEPLKDNCLHSLFHHNPELALNQKNVLLQEETKRMNLMTEIMHPNLIQVVRQSQLSDSEDSEGMDVLGVVGLPREVVRLRLRPIRSSSILHSVEDLSTDSTCVYKASRRKLSRSLYIRPPVTSRLTQSKLADQVIRKEVEIKSEEPEKKPSKGCVEKISRYLDVSLLKSWRFGLLCTSVALMSCGSPYALYYLPAYTVAIGHTKAAAGRLVAISAVLDLCGRLGLGWLCDLHLFCRRKAYIISIFVAGAAVLTLPSLTSWWSLAIVSGAYGLCLGCWFLLVPVLLADAFGTARIASSYGLVRMFQSVAAVSVPPTAGYIRDLTGGFSWCFYGMGSCMVLGTIPVIAFHVSTKNDDNGSTVD
Summary
Uniprot
H9IWQ9
A0A2H1VDB9
A0A0N1PFI6
A0A2A4JPT8
A0A026WLH4
A0A088A6H0
+ More
A0A3L8E1I5 V9IB53 A0A2W1BD54 A0A195CB00 F4WJ94 A0A195BV82 A0A158NEC1 A0A151WT65 A0A151J6K3 A0A195FK71 A0A0M8ZRK7 E1ZZB4 E2B3J9 A0A154PJB9 E9J1H2 A0A1B0CJQ1 W8AK44 B3NSC3 B4P604 B4HNT8 Q5U154 A0A0J9RBJ0 A0A1A9X3B6 A0A1B0FPQ6 A0A1B0BAM3 A0A1I8NS55 A0A1A9V3B0 A0A1B0D651 A0A1B0A4K8 A0A1A9XFE5 A0A1I8MLB9 A0A0M4EKY9 Q28XC0 A0A2A3E3V3 B4H5A3 A0A3B0J5F0 A0A0R3NQ07 B4LIW5 B4N5P8 B4JVS4 A0A336LT44 A0A0J7KU72 E9FRJ5 A0A0P5LQB7 B4QBQ2 A0A0L0BSG6 T1GAW3
A0A3L8E1I5 V9IB53 A0A2W1BD54 A0A195CB00 F4WJ94 A0A195BV82 A0A158NEC1 A0A151WT65 A0A151J6K3 A0A195FK71 A0A0M8ZRK7 E1ZZB4 E2B3J9 A0A154PJB9 E9J1H2 A0A1B0CJQ1 W8AK44 B3NSC3 B4P604 B4HNT8 Q5U154 A0A0J9RBJ0 A0A1A9X3B6 A0A1B0FPQ6 A0A1B0BAM3 A0A1I8NS55 A0A1A9V3B0 A0A1B0D651 A0A1B0A4K8 A0A1A9XFE5 A0A1I8MLB9 A0A0M4EKY9 Q28XC0 A0A2A3E3V3 B4H5A3 A0A3B0J5F0 A0A0R3NQ07 B4LIW5 B4N5P8 B4JVS4 A0A336LT44 A0A0J7KU72 E9FRJ5 A0A0P5LQB7 B4QBQ2 A0A0L0BSG6 T1GAW3
Pubmed
EMBL
BABH01020984
BABH01020985
ODYU01001754
SOQ38392.1
KQ458725
KPJ05376.1
+ More
NWSH01000873 PCG73776.1 KK107154 EZA56895.1 QOIP01000001 RLU26486.1 JR038565 AEY58320.1 KZ150196 PZC72331.1 KQ978023 KYM97880.1 GL888182 EGI65672.1 KQ976401 KYM92534.1 ADTU01013243 ADTU01013244 KQ982769 KYQ50815.1 KQ979851 KYN18838.1 KQ981490 KYN41065.1 KQ435922 KOX68536.1 GL435311 EFN73529.1 GL445346 EFN89751.1 KQ434936 KZC11903.1 GL767671 EFZ13317.1 AJWK01014832 AJWK01014833 GAMC01017571 GAMC01017570 JAB88985.1 CH954179 EDV56425.1 CM000158 EDW90879.1 CH480816 EDW47453.1 AE013599 BT016038 AAF58616.2 AAV36923.1 CM002911 KMY93044.1 CCAG010016854 JXJN01011071 JXJN01011072 JXJN01011073 AJVK01011963 CP012524 ALC42142.1 CM000071 EAL26396.5 KZ288405 PBC26174.1 CH479210 EDW32939.1 OUUW01000001 SPP74973.1 KRT03076.1 CH940648 EDW60415.1 KRF79404.1 KRF79405.1 KRF79406.1 KRF79407.1 KRF79408.1 KRF79409.1 CH964154 EDW79687.1 CH916375 EDV98062.1 UFQT01000176 SSX21164.1 LBMM01003188 KMQ93836.1 GL732523 EFX90175.1 GDIQ01166720 JAK85005.1 CM000362 EDX06669.1 JRES01001425 KNC22987.1 CAQQ02055037 CAQQ02055038
NWSH01000873 PCG73776.1 KK107154 EZA56895.1 QOIP01000001 RLU26486.1 JR038565 AEY58320.1 KZ150196 PZC72331.1 KQ978023 KYM97880.1 GL888182 EGI65672.1 KQ976401 KYM92534.1 ADTU01013243 ADTU01013244 KQ982769 KYQ50815.1 KQ979851 KYN18838.1 KQ981490 KYN41065.1 KQ435922 KOX68536.1 GL435311 EFN73529.1 GL445346 EFN89751.1 KQ434936 KZC11903.1 GL767671 EFZ13317.1 AJWK01014832 AJWK01014833 GAMC01017571 GAMC01017570 JAB88985.1 CH954179 EDV56425.1 CM000158 EDW90879.1 CH480816 EDW47453.1 AE013599 BT016038 AAF58616.2 AAV36923.1 CM002911 KMY93044.1 CCAG010016854 JXJN01011071 JXJN01011072 JXJN01011073 AJVK01011963 CP012524 ALC42142.1 CM000071 EAL26396.5 KZ288405 PBC26174.1 CH479210 EDW32939.1 OUUW01000001 SPP74973.1 KRT03076.1 CH940648 EDW60415.1 KRF79404.1 KRF79405.1 KRF79406.1 KRF79407.1 KRF79408.1 KRF79409.1 CH964154 EDW79687.1 CH916375 EDV98062.1 UFQT01000176 SSX21164.1 LBMM01003188 KMQ93836.1 GL732523 EFX90175.1 GDIQ01166720 JAK85005.1 CM000362 EDX06669.1 JRES01001425 KNC22987.1 CAQQ02055037 CAQQ02055038
Proteomes
UP000005204
UP000053268
UP000218220
UP000053097
UP000005203
UP000279307
+ More
UP000078542 UP000007755 UP000078540 UP000005205 UP000075809 UP000078492 UP000078541 UP000053105 UP000000311 UP000008237 UP000076502 UP000092461 UP000008711 UP000002282 UP000001292 UP000000803 UP000091820 UP000092444 UP000092460 UP000095300 UP000078200 UP000092462 UP000092445 UP000092443 UP000095301 UP000092553 UP000001819 UP000242457 UP000008744 UP000268350 UP000008792 UP000007798 UP000001070 UP000036403 UP000000305 UP000000304 UP000037069 UP000015102
UP000078542 UP000007755 UP000078540 UP000005205 UP000075809 UP000078492 UP000078541 UP000053105 UP000000311 UP000008237 UP000076502 UP000092461 UP000008711 UP000002282 UP000001292 UP000000803 UP000091820 UP000092444 UP000092460 UP000095300 UP000078200 UP000092462 UP000092445 UP000092443 UP000095301 UP000092553 UP000001819 UP000242457 UP000008744 UP000268350 UP000008792 UP000007798 UP000001070 UP000036403 UP000000305 UP000000304 UP000037069 UP000015102
Pfam
PF07690 MFS_1
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9IWQ9
A0A2H1VDB9
A0A0N1PFI6
A0A2A4JPT8
A0A026WLH4
A0A088A6H0
+ More
A0A3L8E1I5 V9IB53 A0A2W1BD54 A0A195CB00 F4WJ94 A0A195BV82 A0A158NEC1 A0A151WT65 A0A151J6K3 A0A195FK71 A0A0M8ZRK7 E1ZZB4 E2B3J9 A0A154PJB9 E9J1H2 A0A1B0CJQ1 W8AK44 B3NSC3 B4P604 B4HNT8 Q5U154 A0A0J9RBJ0 A0A1A9X3B6 A0A1B0FPQ6 A0A1B0BAM3 A0A1I8NS55 A0A1A9V3B0 A0A1B0D651 A0A1B0A4K8 A0A1A9XFE5 A0A1I8MLB9 A0A0M4EKY9 Q28XC0 A0A2A3E3V3 B4H5A3 A0A3B0J5F0 A0A0R3NQ07 B4LIW5 B4N5P8 B4JVS4 A0A336LT44 A0A0J7KU72 E9FRJ5 A0A0P5LQB7 B4QBQ2 A0A0L0BSG6 T1GAW3
A0A3L8E1I5 V9IB53 A0A2W1BD54 A0A195CB00 F4WJ94 A0A195BV82 A0A158NEC1 A0A151WT65 A0A151J6K3 A0A195FK71 A0A0M8ZRK7 E1ZZB4 E2B3J9 A0A154PJB9 E9J1H2 A0A1B0CJQ1 W8AK44 B3NSC3 B4P604 B4HNT8 Q5U154 A0A0J9RBJ0 A0A1A9X3B6 A0A1B0FPQ6 A0A1B0BAM3 A0A1I8NS55 A0A1A9V3B0 A0A1B0D651 A0A1B0A4K8 A0A1A9XFE5 A0A1I8MLB9 A0A0M4EKY9 Q28XC0 A0A2A3E3V3 B4H5A3 A0A3B0J5F0 A0A0R3NQ07 B4LIW5 B4N5P8 B4JVS4 A0A336LT44 A0A0J7KU72 E9FRJ5 A0A0P5LQB7 B4QBQ2 A0A0L0BSG6 T1GAW3
Ontologies
GO
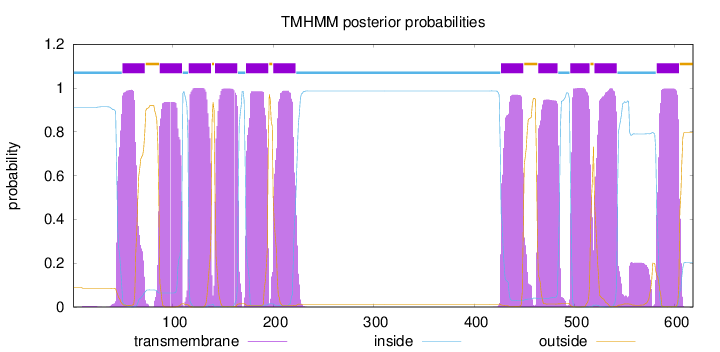
Topology
Length:
618
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
241.68187
Exp number, first 60 AAs:
14.95582
Total prob of N-in:
0.91051
POSSIBLE N-term signal
sequence
inside
1 - 49
TMhelix
50 - 72
outside
73 - 86
TMhelix
87 - 109
inside
110 - 115
TMhelix
116 - 138
outside
139 - 141
TMhelix
142 - 164
inside
165 - 172
TMhelix
173 - 195
outside
196 - 199
TMhelix
200 - 222
inside
223 - 426
TMhelix
427 - 449
outside
450 - 463
TMhelix
464 - 483
inside
484 - 495
TMhelix
496 - 515
outside
516 - 519
TMhelix
520 - 542
inside
543 - 581
TMhelix
582 - 604
outside
605 - 618
Population Genetic Test Statistics
Pi
170.830649
Theta
125.467541
Tajima's D
1.340181
CLR
0.769952
CSRT
0.748912554372281
Interpretation
Uncertain
