Gene
KWMTBOMO06112
Pre Gene Modal
BGIBMGA001692
Annotation
PREDICTED:_intersectin-1_isoform_X3_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.658
Sequence
CDS
ATGATGGCGGACCCGTGGACTATACAAGCACATGAGCATGCGAAGTTCTCGGAGCACTTTAGAAACCTTGGACCTGTCAATGGAAATTTGACGGGCGAACAAGCTAAAAGATTCATGTTACAGTCTCAGCTTCCTCCACCGGTCTTGGGACAAATATGGTCACTGGCGGACACGAACGCCGACGGTAAGCTGGATTTGAAGGAGTTCTCTATAGCTTGCAAGATTATAAACTTGAAACTGCGCGGCTTAGAGGTGCCGAAGATGCTTCCACCATCTCTGATCGCCTCTTTAAGCCCCACAGGAGGCCAATCGCAACAGGTCGTTCCGCCGGCGAAACCAGCCATTCCGCCCCTGCCCAACATACCTCCCCAACCTCTGATCTCCGGTCTATCGCCCACTCAACCGTCCAACCAGTCGTTGCTCGGTGATTTCGGCTCCGGTCAGCCGCTGATCCAACCTTTGGTCCAGCCCGTCAAACCGCCCGTCAGTACGGTGCCGGATCTGATCTCCGGCGTGAAGCCGTTGTCGCAATCCTTGCTAGACTCCCCACCCATGGCCCCCACTGTTCAGCCGTTGATTGGCACTGTCCAGCCATTAATCGGTCCAGATCCACTGATGGGCAGCGTCCAACCCCTGATTGGCTCCCAGCCACTGGCCGGACCCACGCCCCTAGTGAGCTCACCCCAAACTCTAGTCAATTCATCACCATTAGTCTCGTCACAGCCGATTATGTCCAGCCAGCCGTTAATTAGTGCTTCTACGATCTCCGCACAACCTCTTGGAACAATGCAGCCCCTGATAGGGACCGCTCCACCTATTATGGGAGCGACTCAGGCCCTGATCCAGTCTCCCCCCCTGTCTGGTCCGCAACAACCCGCGCCGGTCACGGCCGCGTCGTTTGGGGGTAGTCCCCCGCACCCGGTGGTCGCAGCTTCGGTTCCCGGACAGCCGGTGACGTCTACGCCGATCGCGGCTCTGAACAAAGAGGTCATATCGAAACCAGGATCCGTAGTGACGAGTCCCGAAGTCCCGATGGGAGTGATGAGCCCCCCTCCCATGGAGTGGGGGGTGCCGCAGCCACAGAAACTCAAGTACACGCAGCTGTTCAACGCCACTGACCGCGCGAAGACCGGATCCGTTTCCGGGACGCAGGCGAGGTCCATCATGCTGCAGTCCAGACTCCCACAGCAAAGCCTGGCCCAGATCTGGGCGCTGGCCGATCTGGACGCCGATGGGAAATTGGGCTGCGAAGAGTTCGTCCTGGCGATGTACCTGTGCGAGAAGGCGACGCAGGGCGAGCCGGTGCCCGCCAGGCTGCCTCCGGAACTGATCCCGCCTTCCTTCAGGAGGGAATCGGTTTCGTCCACGAGCAGCGCTAAGTCGTACGAGGAGCGGCGGCGGGAGAACATGGCGCGCGGACAGGCCGAGCTCGAGCGCCGACGGACGCAGCTGGCCGACGCACAGATGAAGGAGAAGGCAGAGAGGGAGAGGAAAGAGCGAGAGGAAGCCGAGAAGAGAGAAAAACTCCGTCTGGAGGCCCAGCAGAGGGAGCAGGAGGAGTTACTCCGTAAACAGAAGGAGGAACAGGAAAAACGCGAAGCTGCACGCAAAGAGATGGAGAGGCAGTGCCAGCTGGAGTGGGAGAAGAAACGGACCAGGGAACTGGAAGCGCTGAGGAGCGAGGAGCAGAGCAAGGTGTTGAGTCTGAAGGCGAAGTGCCAGTCGCTGAGCGCGCAACTCGGCGCCGAGAGCGTCCGCTGCAGCACCCTGGCCGCCAGCATCCGGGACACCAGGACCGCCGTCGCCGCCGACAAGCGGACCATAGACGGCATGAGGTCCGACCGGGACTCCAGCATGGCGGAGATGCAGCAGCTGAAGGCGCGAGTCAAGGAGCTCAACGCGAAGCAGATCGCTTTGAACAAAGAGAAGGCGGAGTTGGACGCCAAGGCTAAAGCCGCAGAGGGCGCTGGTGAGGATGGGAAACTCAGCATGATGGAGGTCACGCTCAAGCAACTCCGGGAGAAGGTGGAGGCCGCCAAAGCTATCGTTGAATCGAAGACCGCGGACCTGCAGCACAACGAGGCCCAGCTGGCGGAGCTGAACGCGACGGTAGCGACGCTCAGCGACAAGTGCGAGAAGGTGTGGAAGCTGTACGAGGAGAAACGGAACGAGTACATCGAGAGGAGGAACACGGCGCCCGCGGAGGGGACCTGGGGCAGCAGCGACGCGGACTGGGGCGCCGCGGACGCCGCGTGGAGCTCCCCCCCGGCCGAGGACGCGTGGGGCGACGCGCCCACCACCACCGCCACGGAGACGGTGCAGACCGCAGCCCCCGGGGGCGAGGGCCCGCAGCCGCCGCGGTGGCGCTGCGTGTACCCGTTCGTGGGGCGCAGCGCCGACGAGCTGAGCGTGCAGCCCGGGGACCTCATCGTGCCGCGCGCGCCCCCCGCCGCGCCAGAGCCCGGCTGGGCCTGGGGCAGCGCCAGGGGTCAGAGCGGCTGGTTCCCGGAATCGTACGTGGAGGACCTCAACGCTCCCACCGCGTTCGTCACCGAGGTCATCGAGACCCTCGAGCCAAAGACGCAGCTTGAAGGGATAGCGGAGGTACCCGAACAGGAGGCCACCAGCGACCTCGGGGGCGCGCTGCCGGCAGTAGACGACGGCGAGTTCTACATAGCGTCGTACCCGTACAGCTCCTCGGAGCCCGGCGACCTGGTGTTCGAGGCCGGCGAGAGGATCGAGGTGGTGCGGCGCGACGGCGACTGGTGGACCGGCAGGATCGGGGCCAGGACCGGCATATTCCCCTCGAACTACGTGTCCCGGGAGGCAGCGCCCGCGGCCACCTCGGAGCCCCAGGACTCCGCCCCGGCCGAGGCGTCCCCCGCACCCCCCGCCGCGCCCGAGCAGGAGGACACGTCGCCCCCCAACACAACTCCGCCGATACCTGCCTCTCAGGACCAGAAGTCCTCGACGCCGATATCGTCAGAGGTGGCCATGATCACGGACCCCGCGCGGCCCCCCTCCGCCGCGGGGGCGCGCCGCGACTCCGGGCGCGACTCCGTCACCGGCCGCCGCAAGCACGAGATAGTGCAGGCGCTCGCCAGCTACACCGCCACCAGCCCGGAGCAGCTGAGTCTGATGAAGGGACAGCTGCTGGTGGTGCGCAAGAAGGCCGACAGCGGCTGGTGGGAGGGCGAGCTGCAGGCCAAGGGCAGGAACCGGCAGGTCGGCTGGTTCCCCGCTTCCTACGTCAAGGTGCTGCAGTCGTCGGGGCGGACCTCGGGCCGCACCACTCCCGTGCTCTCCTCGAAGATGGACGCTCTACCCACGGAGACGGTCATCGACAAGGTGGTGGCGCTGTATCCGTACACGGCTCAAAACGCGGACGAGCTCAGCTTCGAGAAGGACGACATCATAGCCGTCACCGACCGCAGCCAGGACCCGGCGTGGTGGCAAGGCGAACTTCGTGGCATGGTCGGCCTCTTCCCCAGCAATTACGTTACGAAATTAGTTAATTAA
Protein
MMADPWTIQAHEHAKFSEHFRNLGPVNGNLTGEQAKRFMLQSQLPPPVLGQIWSLADTNADGKLDLKEFSIACKIINLKLRGLEVPKMLPPSLIASLSPTGGQSQQVVPPAKPAIPPLPNIPPQPLISGLSPTQPSNQSLLGDFGSGQPLIQPLVQPVKPPVSTVPDLISGVKPLSQSLLDSPPMAPTVQPLIGTVQPLIGPDPLMGSVQPLIGSQPLAGPTPLVSSPQTLVNSSPLVSSQPIMSSQPLISASTISAQPLGTMQPLIGTAPPIMGATQALIQSPPLSGPQQPAPVTAASFGGSPPHPVVAASVPGQPVTSTPIAALNKEVISKPGSVVTSPEVPMGVMSPPPMEWGVPQPQKLKYTQLFNATDRAKTGSVSGTQARSIMLQSRLPQQSLAQIWALADLDADGKLGCEEFVLAMYLCEKATQGEPVPARLPPELIPPSFRRESVSSTSSAKSYEERRRENMARGQAELERRRTQLADAQMKEKAERERKEREEAEKREKLRLEAQQREQEELLRKQKEEQEKREAARKEMERQCQLEWEKKRTRELEALRSEEQSKVLSLKAKCQSLSAQLGAESVRCSTLAASIRDTRTAVAADKRTIDGMRSDRDSSMAEMQQLKARVKELNAKQIALNKEKAELDAKAKAAEGAGEDGKLSMMEVTLKQLREKVEAAKAIVESKTADLQHNEAQLAELNATVATLSDKCEKVWKLYEEKRNEYIERRNTAPAEGTWGSSDADWGAADAAWSSPPAEDAWGDAPTTTATETVQTAAPGGEGPQPPRWRCVYPFVGRSADELSVQPGDLIVPRAPPAAPEPGWAWGSARGQSGWFPESYVEDLNAPTAFVTEVIETLEPKTQLEGIAEVPEQEATSDLGGALPAVDDGEFYIASYPYSSSEPGDLVFEAGERIEVVRRDGDWWTGRIGARTGIFPSNYVSREAAPAATSEPQDSAPAEASPAPPAAPEQEDTSPPNTTPPIPASQDQKSSTPISSEVAMITDPARPPSAAGARRDSGRDSVTGRRKHEIVQALASYTATSPEQLSLMKGQLLVVRKKADSGWWEGELQAKGRNRQVGWFPASYVKVLQSSGRTSGRTTPVLSSKMDALPTETVIDKVVALYPYTAQNADELSFEKDDIIAVTDRSQDPAWWQGELRGMVGLFPSNYVTKLVN
Summary
Uniprot
Pubmed
EMBL
Proteomes
PRIDE
Interpro
IPR001452
SH3_domain
+ More
IPR000261 EH_dom
IPR018247 EF_Hand_1_Ca_BS
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR036028 SH3-like_dom_sf
IPR035979 RBD_domain_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR035899 DBL_dom_sf
IPR000219 DH-domain
IPR001849 PH_domain
IPR035892 C2_domain_sf
IPR000008 C2_dom
IPR011993 PH-like_dom_sf
IPR000261 EH_dom
IPR018247 EF_Hand_1_Ca_BS
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR036028 SH3-like_dom_sf
IPR035979 RBD_domain_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR035899 DBL_dom_sf
IPR000219 DH-domain
IPR001849 PH_domain
IPR035892 C2_domain_sf
IPR000008 C2_dom
IPR011993 PH-like_dom_sf
Gene 3D
ProteinModelPortal
PDB
2KGR
E-value=8.61657e-24,
Score=278
Ontologies
GO
Topology
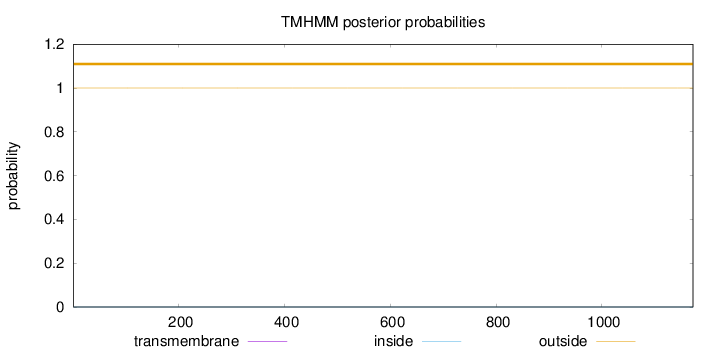
Length:
1172
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00121
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.00003
outside
1 - 1172
Population Genetic Test Statistics
Pi
300.86468
Theta
163.341222
Tajima's D
2.272895
CLR
0.43766
CSRT
0.918454077296135
Interpretation
Uncertain
