Gene
KWMTBOMO06111 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001733
Annotation
PREDICTED:_protein_LSM12_homolog_[Papilio_polytes]
Full name
LSM12 homolog A
Alternative Name
Protein Lsm12
Location in the cell
Cytoplasmic Reliability : 1.04 Mitochondrial Reliability : 1.133 Nuclear Reliability : 1.162
Sequence
CDS
ATGGCGATAGCTAGAGTGATTGACGACGTGTCCTGGGCCGGACAGAGCATCCGAGTATACAATAACAAAATACACCAAGTAATGATCACACCTCCATATAAGGTCGATGACGTCATAGGCGAAAAGGATTCCCAGTCGTATAATTATATTAAGAAATTCGTAGAAAGACACTGGCGCGATCGTCCCACGCCTGCGCCCCAACAATAA
Protein
MAIARVIDDVSWAGQSIRVYNNKIHQVMITPPYKVDDVIGEKDSQSYNYIKKFVERHWRDRPTPAPQQ
Summary
Description
Component of the Atx2-tyf activator complex which functions in the circadian pacemaker neurons to activate the TYF-dependent translation of per and maintain 24 hour periodicity in circadian behaviors. Within the Atx2-tyf complex, likely to function as a molecular adapter which stabilizes the interaction between Atx2 and the translational regulator tyf.
Subunit
Component of the Atx2-tyf activator complex, composed of Atx2, tyf, pAbp, Lsm12. Interacts with tyf, Atx2 and pAbp.
Similarity
Belongs to the LSM12 family.
Keywords
Complete proteome
Reference proteome
Feature
chain LSM12 homolog A
Uniprot
H9IWV0
A0A2W1BEJ6
A0A2A4JRI6
A0A2H1VXJ0
S4P382
I4DPB8
+ More
A0A0N1I7N9 A0A212EXA6 A0A1E1W162 A0A3S2L5V3 U5EQH9 T1E2F8 B0XGZ7 A0A1Q3F9V4 E2AUX3 A0A1B6GS47 A0A0C9PID9 A0A151IFK6 Q170S3 A0A182IVF6 A0A1B0CTY5 A0A0J7L0N4 A0A1B0FY79 A0A195FJL3 A0A151JBJ7 F4WAY7 A0A195BEB0 A0A151XF83 A0A158NRB9 A0A1L8DC82 A0A1L8DCM6 A0A1B6IZ86 E0VNM5 A0A026WFH8 A0A3L8DJV0 A0A1B6KTE3 A0A182QI28 A0A2J7RFB9 A0A182NMW7 E2BLG3 A0A2R7WLN7 A0A1Y1MSF7 D2A635 A0A084VCU0 A0A0T6B4Q7 B3MXT3 V5GUY7 A0A1W7RA99 A0A1D2NMV9 A0A069DRB3 B4PY92 A0A2R5L9V5 B3NXM4 A0A023F8V6 A0A0V0GB73 A0A1W4VWT8 B4R3L1 B4ILA9 Q9VYR0 A0A224XIL0 A0A2A3EBU9 V9IGW0 A0A0L7RCJ7 A0A088AR49 A0A310SH57 A0A1S3CX41 A0A182SNV6 A0A182XWH3 A0A0P4VXZ5 A0A232F9B9 K7IRS4 W5J4V3 A0A182LX51 A0A0P5TWN1 A0A182RFT6 A0A182VYF0 A0A182VEC1 A0A182U4U7 A0A182LQR2 A0A182WZJ8 Q7QK61 A0A182HZP7 A0A182JVG2 A0A0P6AIH0 A0A0A9Z6Z7 A0A0K8T7K6 A0A0P6GET2 A0A226DKV9 A0A182PEA4 A0A0N8B9Y6 C1C052 A0A0P5V7G6 A0A023GIB2 G3MMM4 A0A1E1XU14 A0A023FJ44 A0A1E1WZ46 B4JNB8 A0A023FU34
A0A0N1I7N9 A0A212EXA6 A0A1E1W162 A0A3S2L5V3 U5EQH9 T1E2F8 B0XGZ7 A0A1Q3F9V4 E2AUX3 A0A1B6GS47 A0A0C9PID9 A0A151IFK6 Q170S3 A0A182IVF6 A0A1B0CTY5 A0A0J7L0N4 A0A1B0FY79 A0A195FJL3 A0A151JBJ7 F4WAY7 A0A195BEB0 A0A151XF83 A0A158NRB9 A0A1L8DC82 A0A1L8DCM6 A0A1B6IZ86 E0VNM5 A0A026WFH8 A0A3L8DJV0 A0A1B6KTE3 A0A182QI28 A0A2J7RFB9 A0A182NMW7 E2BLG3 A0A2R7WLN7 A0A1Y1MSF7 D2A635 A0A084VCU0 A0A0T6B4Q7 B3MXT3 V5GUY7 A0A1W7RA99 A0A1D2NMV9 A0A069DRB3 B4PY92 A0A2R5L9V5 B3NXM4 A0A023F8V6 A0A0V0GB73 A0A1W4VWT8 B4R3L1 B4ILA9 Q9VYR0 A0A224XIL0 A0A2A3EBU9 V9IGW0 A0A0L7RCJ7 A0A088AR49 A0A310SH57 A0A1S3CX41 A0A182SNV6 A0A182XWH3 A0A0P4VXZ5 A0A232F9B9 K7IRS4 W5J4V3 A0A182LX51 A0A0P5TWN1 A0A182RFT6 A0A182VYF0 A0A182VEC1 A0A182U4U7 A0A182LQR2 A0A182WZJ8 Q7QK61 A0A182HZP7 A0A182JVG2 A0A0P6AIH0 A0A0A9Z6Z7 A0A0K8T7K6 A0A0P6GET2 A0A226DKV9 A0A182PEA4 A0A0N8B9Y6 C1C052 A0A0P5V7G6 A0A023GIB2 G3MMM4 A0A1E1XU14 A0A023FJ44 A0A1E1WZ46 B4JNB8 A0A023FU34
Pubmed
19121390
28756777
23622113
22651552
26354079
22118469
+ More
24330624 20798317 17510324 21719571 21347285 20566863 24508170 30249741 28004739 18362917 19820115 24438588 17994087 27289101 26334808 17550304 25474469 10731132 12537572 12537569 28388438 25244985 27129103 28648823 20075255 20920257 23761445 20966253 12364791 14747013 17210077 25401762 26823975 30926861 22216098 29209593 28503490
24330624 20798317 17510324 21719571 21347285 20566863 24508170 30249741 28004739 18362917 19820115 24438588 17994087 27289101 26334808 17550304 25474469 10731132 12537572 12537569 28388438 25244985 27129103 28648823 20075255 20920257 23761445 20966253 12364791 14747013 17210077 25401762 26823975 30926861 22216098 29209593 28503490
EMBL
BABH01020981
KZ150158
PZC72751.1
NWSH01000714
PCG74645.1
ODYU01005049
+ More
SOQ45550.1 GAIX01011710 JAA80850.1 AK403517 KQ458725 BAM19758.1 KPJ05362.1 KQ460546 KPJ14050.1 AGBW02011796 OWR46115.1 GDQN01010382 JAT80672.1 RSAL01000131 RVE46410.1 GANO01003258 JAB56613.1 GALA01000759 JAA94093.1 DS233082 EDS27965.1 GFDL01010695 JAV24350.1 GL442898 EFN62781.1 GECZ01004527 JAS65242.1 GBYB01000693 JAG70460.1 KQ977771 KYM99978.1 CH477469 EAT40444.1 EAT40445.1 AJWK01028171 AJWK01028172 LBMM01001417 KMQ96357.1 AJVK01000789 AJVK01000790 AJVK01000791 AJVK01000792 KQ981522 KYN40556.1 KQ979122 KYN22531.1 GL888056 EGI68607.1 KQ976504 KYM82921.1 KQ982205 KYQ58995.1 ADTU01023849 ADTU01023850 GFDF01010013 JAV04071.1 GFDF01010014 JAV04070.1 GECU01015480 JAS92226.1 DS235340 EEB14981.1 KK107238 EZA54683.1 QOIP01000007 RLU20677.1 GEBQ01025306 JAT14671.1 AXCN02001550 NEVH01004413 PNF39533.1 GL449017 EFN83515.1 KK854868 PTY19345.1 GEZM01022992 JAV88623.1 KQ971346 EFA04985.1 ATLV01010866 KE524621 KFB35784.1 LJIG01009825 KRT82306.1 CH902630 EDV38548.1 GALX01000522 JAB67944.1 GFAH01000332 JAV48057.1 LJIJ01000002 ODN06592.1 GBGD01002648 JAC86241.1 CM000162 EDX00965.1 GGLE01002109 MBY06235.1 CH954180 EDV47325.1 GBBI01000857 JAC17855.1 GECL01000762 JAP05362.1 CM000366 EDX17713.1 CH480868 EDW53755.1 AE014298 AY069351 GFTR01004100 JAW12326.1 KZ288289 PBC29253.1 JR043102 AEY59726.1 KQ414615 KOC68525.1 KQ763581 OAD54969.1 GDKW01002136 JAI54459.1 NNAY01000701 OXU26907.1 ADMH02002134 ETN58423.1 AXCM01003899 GDIP01123061 JAL80653.1 AAAB01008799 EAA03679.4 EGK96214.1 APCN01001988 GDIP01030514 JAM73201.1 GBHO01002568 GBHO01002567 GDHC01017555 JAG41036.1 JAG41037.1 JAQ01074.1 GBRD01004318 JAG61503.1 GDIQ01046536 JAN48201.1 LNIX01000017 MH602547 OXA45760.1 QBH72792.1 GDIQ01198371 JAK53354.1 BT080231 ACO14655.1 GDIP01104564 JAL99150.1 GBBM01002650 JAC32768.1 JO843125 AEO34742.1 GFAA01000644 JAU02791.1 GBBK01003814 JAC20668.1 GFAC01006921 JAT92267.1 CH916371 EDV92211.1 GBBL01001989 JAC25331.1
SOQ45550.1 GAIX01011710 JAA80850.1 AK403517 KQ458725 BAM19758.1 KPJ05362.1 KQ460546 KPJ14050.1 AGBW02011796 OWR46115.1 GDQN01010382 JAT80672.1 RSAL01000131 RVE46410.1 GANO01003258 JAB56613.1 GALA01000759 JAA94093.1 DS233082 EDS27965.1 GFDL01010695 JAV24350.1 GL442898 EFN62781.1 GECZ01004527 JAS65242.1 GBYB01000693 JAG70460.1 KQ977771 KYM99978.1 CH477469 EAT40444.1 EAT40445.1 AJWK01028171 AJWK01028172 LBMM01001417 KMQ96357.1 AJVK01000789 AJVK01000790 AJVK01000791 AJVK01000792 KQ981522 KYN40556.1 KQ979122 KYN22531.1 GL888056 EGI68607.1 KQ976504 KYM82921.1 KQ982205 KYQ58995.1 ADTU01023849 ADTU01023850 GFDF01010013 JAV04071.1 GFDF01010014 JAV04070.1 GECU01015480 JAS92226.1 DS235340 EEB14981.1 KK107238 EZA54683.1 QOIP01000007 RLU20677.1 GEBQ01025306 JAT14671.1 AXCN02001550 NEVH01004413 PNF39533.1 GL449017 EFN83515.1 KK854868 PTY19345.1 GEZM01022992 JAV88623.1 KQ971346 EFA04985.1 ATLV01010866 KE524621 KFB35784.1 LJIG01009825 KRT82306.1 CH902630 EDV38548.1 GALX01000522 JAB67944.1 GFAH01000332 JAV48057.1 LJIJ01000002 ODN06592.1 GBGD01002648 JAC86241.1 CM000162 EDX00965.1 GGLE01002109 MBY06235.1 CH954180 EDV47325.1 GBBI01000857 JAC17855.1 GECL01000762 JAP05362.1 CM000366 EDX17713.1 CH480868 EDW53755.1 AE014298 AY069351 GFTR01004100 JAW12326.1 KZ288289 PBC29253.1 JR043102 AEY59726.1 KQ414615 KOC68525.1 KQ763581 OAD54969.1 GDKW01002136 JAI54459.1 NNAY01000701 OXU26907.1 ADMH02002134 ETN58423.1 AXCM01003899 GDIP01123061 JAL80653.1 AAAB01008799 EAA03679.4 EGK96214.1 APCN01001988 GDIP01030514 JAM73201.1 GBHO01002568 GBHO01002567 GDHC01017555 JAG41036.1 JAG41037.1 JAQ01074.1 GBRD01004318 JAG61503.1 GDIQ01046536 JAN48201.1 LNIX01000017 MH602547 OXA45760.1 QBH72792.1 GDIQ01198371 JAK53354.1 BT080231 ACO14655.1 GDIP01104564 JAL99150.1 GBBM01002650 JAC32768.1 JO843125 AEO34742.1 GFAA01000644 JAU02791.1 GBBK01003814 JAC20668.1 GFAC01006921 JAT92267.1 CH916371 EDV92211.1 GBBL01001989 JAC25331.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000283053
+ More
UP000002320 UP000000311 UP000078542 UP000008820 UP000075880 UP000092461 UP000036403 UP000092462 UP000078541 UP000078492 UP000007755 UP000078540 UP000075809 UP000005205 UP000009046 UP000053097 UP000279307 UP000075886 UP000235965 UP000075884 UP000008237 UP000007266 UP000030765 UP000007801 UP000094527 UP000002282 UP000008711 UP000192221 UP000000304 UP000001292 UP000000803 UP000242457 UP000053825 UP000005203 UP000079169 UP000075901 UP000076408 UP000215335 UP000002358 UP000000673 UP000075883 UP000075900 UP000075920 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075881 UP000198287 UP000075885 UP000001070
UP000002320 UP000000311 UP000078542 UP000008820 UP000075880 UP000092461 UP000036403 UP000092462 UP000078541 UP000078492 UP000007755 UP000078540 UP000075809 UP000005205 UP000009046 UP000053097 UP000279307 UP000075886 UP000235965 UP000075884 UP000008237 UP000007266 UP000030765 UP000007801 UP000094527 UP000002282 UP000008711 UP000192221 UP000000304 UP000001292 UP000000803 UP000242457 UP000053825 UP000005203 UP000079169 UP000075901 UP000076408 UP000215335 UP000002358 UP000000673 UP000075883 UP000075900 UP000075920 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075881 UP000198287 UP000075885 UP000001070
Pfam
PF09793 AD
ProteinModelPortal
H9IWV0
A0A2W1BEJ6
A0A2A4JRI6
A0A2H1VXJ0
S4P382
I4DPB8
+ More
A0A0N1I7N9 A0A212EXA6 A0A1E1W162 A0A3S2L5V3 U5EQH9 T1E2F8 B0XGZ7 A0A1Q3F9V4 E2AUX3 A0A1B6GS47 A0A0C9PID9 A0A151IFK6 Q170S3 A0A182IVF6 A0A1B0CTY5 A0A0J7L0N4 A0A1B0FY79 A0A195FJL3 A0A151JBJ7 F4WAY7 A0A195BEB0 A0A151XF83 A0A158NRB9 A0A1L8DC82 A0A1L8DCM6 A0A1B6IZ86 E0VNM5 A0A026WFH8 A0A3L8DJV0 A0A1B6KTE3 A0A182QI28 A0A2J7RFB9 A0A182NMW7 E2BLG3 A0A2R7WLN7 A0A1Y1MSF7 D2A635 A0A084VCU0 A0A0T6B4Q7 B3MXT3 V5GUY7 A0A1W7RA99 A0A1D2NMV9 A0A069DRB3 B4PY92 A0A2R5L9V5 B3NXM4 A0A023F8V6 A0A0V0GB73 A0A1W4VWT8 B4R3L1 B4ILA9 Q9VYR0 A0A224XIL0 A0A2A3EBU9 V9IGW0 A0A0L7RCJ7 A0A088AR49 A0A310SH57 A0A1S3CX41 A0A182SNV6 A0A182XWH3 A0A0P4VXZ5 A0A232F9B9 K7IRS4 W5J4V3 A0A182LX51 A0A0P5TWN1 A0A182RFT6 A0A182VYF0 A0A182VEC1 A0A182U4U7 A0A182LQR2 A0A182WZJ8 Q7QK61 A0A182HZP7 A0A182JVG2 A0A0P6AIH0 A0A0A9Z6Z7 A0A0K8T7K6 A0A0P6GET2 A0A226DKV9 A0A182PEA4 A0A0N8B9Y6 C1C052 A0A0P5V7G6 A0A023GIB2 G3MMM4 A0A1E1XU14 A0A023FJ44 A0A1E1WZ46 B4JNB8 A0A023FU34
A0A0N1I7N9 A0A212EXA6 A0A1E1W162 A0A3S2L5V3 U5EQH9 T1E2F8 B0XGZ7 A0A1Q3F9V4 E2AUX3 A0A1B6GS47 A0A0C9PID9 A0A151IFK6 Q170S3 A0A182IVF6 A0A1B0CTY5 A0A0J7L0N4 A0A1B0FY79 A0A195FJL3 A0A151JBJ7 F4WAY7 A0A195BEB0 A0A151XF83 A0A158NRB9 A0A1L8DC82 A0A1L8DCM6 A0A1B6IZ86 E0VNM5 A0A026WFH8 A0A3L8DJV0 A0A1B6KTE3 A0A182QI28 A0A2J7RFB9 A0A182NMW7 E2BLG3 A0A2R7WLN7 A0A1Y1MSF7 D2A635 A0A084VCU0 A0A0T6B4Q7 B3MXT3 V5GUY7 A0A1W7RA99 A0A1D2NMV9 A0A069DRB3 B4PY92 A0A2R5L9V5 B3NXM4 A0A023F8V6 A0A0V0GB73 A0A1W4VWT8 B4R3L1 B4ILA9 Q9VYR0 A0A224XIL0 A0A2A3EBU9 V9IGW0 A0A0L7RCJ7 A0A088AR49 A0A310SH57 A0A1S3CX41 A0A182SNV6 A0A182XWH3 A0A0P4VXZ5 A0A232F9B9 K7IRS4 W5J4V3 A0A182LX51 A0A0P5TWN1 A0A182RFT6 A0A182VYF0 A0A182VEC1 A0A182U4U7 A0A182LQR2 A0A182WZJ8 Q7QK61 A0A182HZP7 A0A182JVG2 A0A0P6AIH0 A0A0A9Z6Z7 A0A0K8T7K6 A0A0P6GET2 A0A226DKV9 A0A182PEA4 A0A0N8B9Y6 C1C052 A0A0P5V7G6 A0A023GIB2 G3MMM4 A0A1E1XU14 A0A023FJ44 A0A1E1WZ46 B4JNB8 A0A023FU34
Ontologies
PANTHER
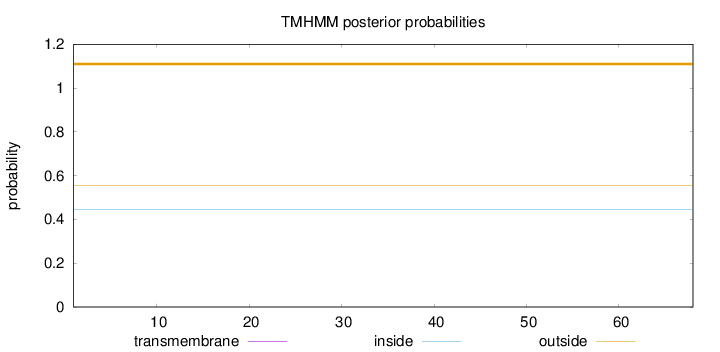
Topology
Length:
68
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.44547
outside
1 - 68
Population Genetic Test Statistics
Pi
156.922388
Theta
182.67833
Tajima's D
2.876666
CLR
0.355662
CSRT
0.970551472426379
Interpretation
Uncertain
