Pre Gene Modal
BGIBMGA001731
Annotation
PREDICTED:_CDKN2A-interacting_protein-like_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 3.047
Sequence
CDS
ATGAAACCTTCTATAGACGTAGATTCGTTGAGAACGGAGCACGAATCTGAGGAACAGTGGGAAGTTAGACGTAGTTTCCTTTTAGAACACAAAGACGATTTTCCAGAAGCTGAATTAGTGACACTGGCGCAAATCTATACCAATATCGAATTTTTAGGATGCAGATATCCACCTCAAACAATGAAACGCATTTCGGTATTGGCCGAGAAAGTGTCCGCCAAGTACAAGGAGAGCAGAAAGAATAAGTTGAAACGAACTTTCGTGCAAGCCAGCGATGCGGCCGAGCAAAAAGCTAAACGTACCTTCAAGAAATAG
Protein
MKPSIDVDSLRTEHESEEQWEVRRSFLLEHKDDFPEAELVTLAQIYTNIEFLGCRYPPQTMKRISVLAEKVSAKYKESRKNKLKRTFVQASDAAEQKAKRTFKK
Summary
Uniprot
H9IWU8
S4P431
A0A2W1BIU8
A0A0N0PCH8
A0A212F2S8
A0A2A4JTZ6
+ More
A0A2H1VNE1 I4DPB7 A0A0L7LP85 A0A1E1WK47 A0A1E1WUN2 A0A1E1WMU5 A0A1E1WVE5 A0A3S2NB42 A0A067QWE1 A0A2J7PZY6 A0A1B0DCU5 V9I9G1 A0A0L7QUE0 A0A3R7PTP9 A0A154PRB4 A0A088A109 A0A2S2Q9W2 A0A2A4JSZ1 A0A182YDW3 A0A1B6JU41 A0A034WSR3 B4QYI8 B3P0V4 B4HM27 B4PS44 Q9VF83 B3MSU5 A0A1Q3FBP5 A0A1B0CJ31 A0A2M4AWU0 E2BVT2 B4M5Z9 A0A1W4VGB0 U5EQB1 E9J4R2 B4KAQ5 A0A226DBV3 A0A0R3NKS7 B4GNU4 Q29BE6 A0A151II79 W8C2V1 A0A0L0CJJ1 B4JU46 A0A1B6H1D1 A0A2M3ZJL1 A0A1J1I3N5 A0A0A1WSM1 A0A3L8D3A2 A0A232FAC7 A0A0M5J0C3 K7JAE8 B0WFV5 A0A0P4W2U1 E2AX34 A0A182FLX1 A0A2M4C4D0 A0A023ECV5 A0A023ECU1 A0A0K8V390 A0A034WR47 A0A3B0K537 W5JER4 F4X4R9 A0A1Q3F5S3 A0A151HY18 A0A158NWB8 A0A1Q3F4D7 A0A195ET28 A0A151WX27 A0A2H1WHM0 N6UEI5 A0A182GMR1 A0A1E1WJ10 A0A2M4BQQ2 E0VTP8 A0A2H8TEP5 C4WVP8 A0A1B6J0J5 U4UB57 A0A2S2NV77 A0A336LIR6 A0A194PSR3 A0A0P4W4W5 A0A3R7QPM8 A0A1Z1MZS7 B0WCX1 A0A1S4FJE6 Q16YV8 A0A2S2Q2W6 A0A1W4X0D5 V5I6B8 A0A3R7SVA4 A0A1B0AL29
A0A2H1VNE1 I4DPB7 A0A0L7LP85 A0A1E1WK47 A0A1E1WUN2 A0A1E1WMU5 A0A1E1WVE5 A0A3S2NB42 A0A067QWE1 A0A2J7PZY6 A0A1B0DCU5 V9I9G1 A0A0L7QUE0 A0A3R7PTP9 A0A154PRB4 A0A088A109 A0A2S2Q9W2 A0A2A4JSZ1 A0A182YDW3 A0A1B6JU41 A0A034WSR3 B4QYI8 B3P0V4 B4HM27 B4PS44 Q9VF83 B3MSU5 A0A1Q3FBP5 A0A1B0CJ31 A0A2M4AWU0 E2BVT2 B4M5Z9 A0A1W4VGB0 U5EQB1 E9J4R2 B4KAQ5 A0A226DBV3 A0A0R3NKS7 B4GNU4 Q29BE6 A0A151II79 W8C2V1 A0A0L0CJJ1 B4JU46 A0A1B6H1D1 A0A2M3ZJL1 A0A1J1I3N5 A0A0A1WSM1 A0A3L8D3A2 A0A232FAC7 A0A0M5J0C3 K7JAE8 B0WFV5 A0A0P4W2U1 E2AX34 A0A182FLX1 A0A2M4C4D0 A0A023ECV5 A0A023ECU1 A0A0K8V390 A0A034WR47 A0A3B0K537 W5JER4 F4X4R9 A0A1Q3F5S3 A0A151HY18 A0A158NWB8 A0A1Q3F4D7 A0A195ET28 A0A151WX27 A0A2H1WHM0 N6UEI5 A0A182GMR1 A0A1E1WJ10 A0A2M4BQQ2 E0VTP8 A0A2H8TEP5 C4WVP8 A0A1B6J0J5 U4UB57 A0A2S2NV77 A0A336LIR6 A0A194PSR3 A0A0P4W4W5 A0A3R7QPM8 A0A1Z1MZS7 B0WCX1 A0A1S4FJE6 Q16YV8 A0A2S2Q2W6 A0A1W4X0D5 V5I6B8 A0A3R7SVA4 A0A1B0AL29
Pubmed
19121390
23622113
28756777
26354079
22118469
22651552
+ More
26227816 24845553 25244985 25348373 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 21282665 15632085 24495485 26108605 25830018 30249741 28648823 20075255 24945155 20920257 23761445 21719571 21347285 23537049 26483478 20566863 28564581 17510324
26227816 24845553 25244985 25348373 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 21282665 15632085 24495485 26108605 25830018 30249741 28648823 20075255 24945155 20920257 23761445 21719571 21347285 23537049 26483478 20566863 28564581 17510324
EMBL
BABH01020978
GAIX01009222
JAA83338.1
KZ150158
PZC72756.1
KQ460546
+ More
KPJ14054.1 AGBW02010691 OWR48032.1 NWSH01000615 PCG75289.1 ODYU01003490 SOQ42355.1 AK403516 KQ458725 BAM19757.1 KPJ05359.1 JTDY01000440 KOB77159.1 GDQN01003671 JAT87383.1 GDQN01000324 JAT90730.1 GDQN01002786 JAT88268.1 GDQN01000110 JAT90944.1 RSAL01000131 RVE46407.1 KK852869 KDR14653.1 NEVH01020330 PNF21896.1 AJVK01031385 JR037382 JR037384 AEY57747.1 AEY57748.1 KQ414735 KOC62277.1 QCYY01001609 ROT76786.1 KQ435078 KZC14465.1 GGMS01005325 MBY74528.1 NWSH01000684 PCG74808.1 GECU01004979 JAT02728.1 GAKP01002139 JAC56813.1 CM000364 EDX12833.1 CH954181 EDV49002.1 CH480815 EDW42064.1 CM000160 EDW97464.1 AE014297 AY051823 AAF55177.2 AAK93247.1 CH902623 EDV30335.1 GFDL01010127 JAV24918.1 AJWK01013988 GGFK01011851 MBW45172.1 GL450957 EFN80210.1 CH940652 EDW59075.2 GANO01004372 JAB55499.1 GL768112 EFZ12188.1 CH933806 EDW16792.1 LNIX01000025 OXA42610.1 CM000070 KRS99746.1 CH479186 EDW38827.1 EAL27052.2 KQ977505 KYN02266.1 GAMC01006374 JAC00182.1 JRES01000310 KNC32410.1 CH916374 EDV91016.1 GECZ01001277 JAS68492.1 GGFM01007960 MBW28711.1 CVRI01000040 CRK94951.1 GBXI01012646 JAD01646.1 QOIP01000014 RLU14982.1 NNAY01000604 OXU27430.1 CP012526 ALC47486.1 AAZX01006898 DS231921 EDS26490.1 GDRN01089471 JAI60654.1 GL443520 EFN61984.1 GGFJ01011046 MBW60187.1 GAPW01006398 GAPW01006397 GAPW01006396 GAPW01006394 GAPW01006393 JAC07202.1 GAPW01006395 JAC07203.1 GDHF01019056 GDHF01011369 JAI33258.1 JAI40945.1 GAKP01002140 JAC56812.1 OUUW01000005 SPP80736.1 ADMH02001606 ETN61833.1 GL888679 EGI58492.1 GFDL01012131 JAV22914.1 KQ976730 KYM76461.1 ADTU01027870 GFDL01012652 JAV22393.1 KQ981993 KYN31067.1 KQ982669 KYQ52464.1 ODYU01008721 SOQ52565.1 APGK01025445 APGK01025446 KB740591 ENN80150.1 JXUM01075013 KQ562863 KXJ74982.1 GDQN01004080 JAT86974.1 GGFJ01006193 MBW55334.1 DS235769 EEB16754.1 GFXV01000772 MBW12577.1 ABLF02013593 AK341614 BAH71968.1 GECU01015001 JAS92705.1 KB632226 ERL90292.1 GGMR01008333 MBY20952.1 UFQT01000015 SSX17730.1 KQ459593 KPI96471.1 GDRN01089472 JAI60653.1 QCYY01000160 ROT85874.1 KY864366 ARW71688.1 DS231891 EDS43931.1 CH477508 EAT39803.1 GGMS01002872 MBY72075.1 GALX01008460 JAB60006.1 ROT76788.1 JXJN01029743
KPJ14054.1 AGBW02010691 OWR48032.1 NWSH01000615 PCG75289.1 ODYU01003490 SOQ42355.1 AK403516 KQ458725 BAM19757.1 KPJ05359.1 JTDY01000440 KOB77159.1 GDQN01003671 JAT87383.1 GDQN01000324 JAT90730.1 GDQN01002786 JAT88268.1 GDQN01000110 JAT90944.1 RSAL01000131 RVE46407.1 KK852869 KDR14653.1 NEVH01020330 PNF21896.1 AJVK01031385 JR037382 JR037384 AEY57747.1 AEY57748.1 KQ414735 KOC62277.1 QCYY01001609 ROT76786.1 KQ435078 KZC14465.1 GGMS01005325 MBY74528.1 NWSH01000684 PCG74808.1 GECU01004979 JAT02728.1 GAKP01002139 JAC56813.1 CM000364 EDX12833.1 CH954181 EDV49002.1 CH480815 EDW42064.1 CM000160 EDW97464.1 AE014297 AY051823 AAF55177.2 AAK93247.1 CH902623 EDV30335.1 GFDL01010127 JAV24918.1 AJWK01013988 GGFK01011851 MBW45172.1 GL450957 EFN80210.1 CH940652 EDW59075.2 GANO01004372 JAB55499.1 GL768112 EFZ12188.1 CH933806 EDW16792.1 LNIX01000025 OXA42610.1 CM000070 KRS99746.1 CH479186 EDW38827.1 EAL27052.2 KQ977505 KYN02266.1 GAMC01006374 JAC00182.1 JRES01000310 KNC32410.1 CH916374 EDV91016.1 GECZ01001277 JAS68492.1 GGFM01007960 MBW28711.1 CVRI01000040 CRK94951.1 GBXI01012646 JAD01646.1 QOIP01000014 RLU14982.1 NNAY01000604 OXU27430.1 CP012526 ALC47486.1 AAZX01006898 DS231921 EDS26490.1 GDRN01089471 JAI60654.1 GL443520 EFN61984.1 GGFJ01011046 MBW60187.1 GAPW01006398 GAPW01006397 GAPW01006396 GAPW01006394 GAPW01006393 JAC07202.1 GAPW01006395 JAC07203.1 GDHF01019056 GDHF01011369 JAI33258.1 JAI40945.1 GAKP01002140 JAC56812.1 OUUW01000005 SPP80736.1 ADMH02001606 ETN61833.1 GL888679 EGI58492.1 GFDL01012131 JAV22914.1 KQ976730 KYM76461.1 ADTU01027870 GFDL01012652 JAV22393.1 KQ981993 KYN31067.1 KQ982669 KYQ52464.1 ODYU01008721 SOQ52565.1 APGK01025445 APGK01025446 KB740591 ENN80150.1 JXUM01075013 KQ562863 KXJ74982.1 GDQN01004080 JAT86974.1 GGFJ01006193 MBW55334.1 DS235769 EEB16754.1 GFXV01000772 MBW12577.1 ABLF02013593 AK341614 BAH71968.1 GECU01015001 JAS92705.1 KB632226 ERL90292.1 GGMR01008333 MBY20952.1 UFQT01000015 SSX17730.1 KQ459593 KPI96471.1 GDRN01089472 JAI60653.1 QCYY01000160 ROT85874.1 KY864366 ARW71688.1 DS231891 EDS43931.1 CH477508 EAT39803.1 GGMS01002872 MBY72075.1 GALX01008460 JAB60006.1 ROT76788.1 JXJN01029743
Proteomes
UP000005204
UP000053240
UP000007151
UP000218220
UP000053268
UP000037510
+ More
UP000283053 UP000027135 UP000235965 UP000092462 UP000053825 UP000283509 UP000076502 UP000005203 UP000076408 UP000000304 UP000008711 UP000001292 UP000002282 UP000000803 UP000007801 UP000092461 UP000008237 UP000008792 UP000192221 UP000009192 UP000198287 UP000001819 UP000008744 UP000078542 UP000037069 UP000001070 UP000183832 UP000279307 UP000215335 UP000092553 UP000002358 UP000002320 UP000000311 UP000069272 UP000268350 UP000000673 UP000007755 UP000078540 UP000005205 UP000078541 UP000075809 UP000019118 UP000069940 UP000249989 UP000009046 UP000007819 UP000030742 UP000008820 UP000192223 UP000092460
UP000283053 UP000027135 UP000235965 UP000092462 UP000053825 UP000283509 UP000076502 UP000005203 UP000076408 UP000000304 UP000008711 UP000001292 UP000002282 UP000000803 UP000007801 UP000092461 UP000008237 UP000008792 UP000192221 UP000009192 UP000198287 UP000001819 UP000008744 UP000078542 UP000037069 UP000001070 UP000183832 UP000279307 UP000215335 UP000092553 UP000002358 UP000002320 UP000000311 UP000069272 UP000268350 UP000000673 UP000007755 UP000078540 UP000005205 UP000078541 UP000075809 UP000019118 UP000069940 UP000249989 UP000009046 UP000007819 UP000030742 UP000008820 UP000192223 UP000092460
Interpro
Gene 3D
ProteinModelPortal
H9IWU8
S4P431
A0A2W1BIU8
A0A0N0PCH8
A0A212F2S8
A0A2A4JTZ6
+ More
A0A2H1VNE1 I4DPB7 A0A0L7LP85 A0A1E1WK47 A0A1E1WUN2 A0A1E1WMU5 A0A1E1WVE5 A0A3S2NB42 A0A067QWE1 A0A2J7PZY6 A0A1B0DCU5 V9I9G1 A0A0L7QUE0 A0A3R7PTP9 A0A154PRB4 A0A088A109 A0A2S2Q9W2 A0A2A4JSZ1 A0A182YDW3 A0A1B6JU41 A0A034WSR3 B4QYI8 B3P0V4 B4HM27 B4PS44 Q9VF83 B3MSU5 A0A1Q3FBP5 A0A1B0CJ31 A0A2M4AWU0 E2BVT2 B4M5Z9 A0A1W4VGB0 U5EQB1 E9J4R2 B4KAQ5 A0A226DBV3 A0A0R3NKS7 B4GNU4 Q29BE6 A0A151II79 W8C2V1 A0A0L0CJJ1 B4JU46 A0A1B6H1D1 A0A2M3ZJL1 A0A1J1I3N5 A0A0A1WSM1 A0A3L8D3A2 A0A232FAC7 A0A0M5J0C3 K7JAE8 B0WFV5 A0A0P4W2U1 E2AX34 A0A182FLX1 A0A2M4C4D0 A0A023ECV5 A0A023ECU1 A0A0K8V390 A0A034WR47 A0A3B0K537 W5JER4 F4X4R9 A0A1Q3F5S3 A0A151HY18 A0A158NWB8 A0A1Q3F4D7 A0A195ET28 A0A151WX27 A0A2H1WHM0 N6UEI5 A0A182GMR1 A0A1E1WJ10 A0A2M4BQQ2 E0VTP8 A0A2H8TEP5 C4WVP8 A0A1B6J0J5 U4UB57 A0A2S2NV77 A0A336LIR6 A0A194PSR3 A0A0P4W4W5 A0A3R7QPM8 A0A1Z1MZS7 B0WCX1 A0A1S4FJE6 Q16YV8 A0A2S2Q2W6 A0A1W4X0D5 V5I6B8 A0A3R7SVA4 A0A1B0AL29
A0A2H1VNE1 I4DPB7 A0A0L7LP85 A0A1E1WK47 A0A1E1WUN2 A0A1E1WMU5 A0A1E1WVE5 A0A3S2NB42 A0A067QWE1 A0A2J7PZY6 A0A1B0DCU5 V9I9G1 A0A0L7QUE0 A0A3R7PTP9 A0A154PRB4 A0A088A109 A0A2S2Q9W2 A0A2A4JSZ1 A0A182YDW3 A0A1B6JU41 A0A034WSR3 B4QYI8 B3P0V4 B4HM27 B4PS44 Q9VF83 B3MSU5 A0A1Q3FBP5 A0A1B0CJ31 A0A2M4AWU0 E2BVT2 B4M5Z9 A0A1W4VGB0 U5EQB1 E9J4R2 B4KAQ5 A0A226DBV3 A0A0R3NKS7 B4GNU4 Q29BE6 A0A151II79 W8C2V1 A0A0L0CJJ1 B4JU46 A0A1B6H1D1 A0A2M3ZJL1 A0A1J1I3N5 A0A0A1WSM1 A0A3L8D3A2 A0A232FAC7 A0A0M5J0C3 K7JAE8 B0WFV5 A0A0P4W2U1 E2AX34 A0A182FLX1 A0A2M4C4D0 A0A023ECV5 A0A023ECU1 A0A0K8V390 A0A034WR47 A0A3B0K537 W5JER4 F4X4R9 A0A1Q3F5S3 A0A151HY18 A0A158NWB8 A0A1Q3F4D7 A0A195ET28 A0A151WX27 A0A2H1WHM0 N6UEI5 A0A182GMR1 A0A1E1WJ10 A0A2M4BQQ2 E0VTP8 A0A2H8TEP5 C4WVP8 A0A1B6J0J5 U4UB57 A0A2S2NV77 A0A336LIR6 A0A194PSR3 A0A0P4W4W5 A0A3R7QPM8 A0A1Z1MZS7 B0WCX1 A0A1S4FJE6 Q16YV8 A0A2S2Q2W6 A0A1W4X0D5 V5I6B8 A0A3R7SVA4 A0A1B0AL29
PDB
5FIR
E-value=3.00683e-10,
Score=149
Ontologies
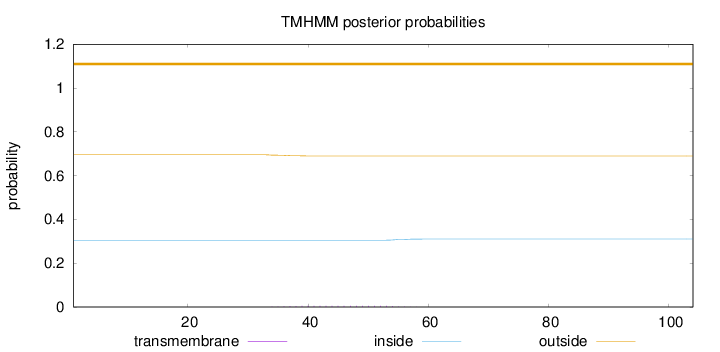
Topology
Length:
104
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10808
Exp number, first 60 AAs:
0.10808
Total prob of N-in:
0.30473
outside
1 - 104
Population Genetic Test Statistics
Pi
160.667639
Theta
78.105336
Tajima's D
2.084796
CLR
15.519634
CSRT
0.896555172241388
Interpretation
Uncertain
