Gene
KWMTBOMO06106 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001695
Annotation
PREDICTED:_lysosomal_alpha-mannosidase-like?_partial_[Bombyx_mori]
Full name
Alpha-mannosidase
Location in the cell
Cytoplasmic Reliability : 2.544
Sequence
CDS
ATGCTGGTACTCGCGCTGCTGTGCCTCCTGAGCATATCATCAAGCGTTCCCCTGCATGACCAGATAACTCCGAGGCCCGGGGAGACGTGTGGCTATGAGAGCTGTCCGAAGACCGAGCCGGGTGTGCTGAACGTCCACATCGTACCCCACACCCACGACGACGTCGGCTGGCTGAAGACCGTCGACCAGTACTATTACGGAAGCAGGAACAACATACAGAAGGCTGGCGTTCAATACATTCTGGATTCGGTTGTCAAGGAGCTGTGGGAAGACCCGAAGCGACGGTTTATCTACGTGGAGACGGCTTTCTTCTGGAAGTGGTGGGTCAGGCAGTCCCGTCGGGTCAAGAGTAAGGTCCACACCCTCGTCAGGCAGGGGCGGCTTCAATTCGTCGGTGGGGCGTGGTCCATGAACGATGAGGCCGCAGTGCACTATCAAAGTACCATCGATCAATTCACTTGGGGTCTCAGCAAATTGAACGAGACCTTCGGTTACTGCGGCTTGCCCCGGGTTGGCTGGCAGATCGACCCGTTCGGGCACTCGAGGGAGCTGGCTTCGCTGCTGTCCGCCATGGGATATGACGGCTTGTTCCTGGGCAGGATCGATTATCAAGACAAGAGGACCAGACTCCGAGACAAGACCATGGAGATGGTATGGAGGGGCGACGACGACTTGGGCAAGAAGTCAGACATCTTCACGGGCGCTTTGTACAACACATACTCTCCGCCGGCCGGGTTCTGCTTCGACGTCCTGTGTGACGACGAGCCCATAGTGGACGACGCTGGCTCGCCGCTGTACAATGTCGACAACAAGATGACCCTATTTCTGAACTACGTTAAAAAGCAATCGCAACATTATCGAACGGACAATGTCATCATCACGATGGGAGGAGATTTCACTTACCAAGACGCCGCGATGTGGTACACGAATCTGGACAAGCTGATTGAATATTCAAATCTGAAGGCGGCCAAAGACGATCTGAAGATTAAGCTGTTTTATTCGACGCCTAATTGTTATCTGAAAGCCATAAAAGACGCGAATCCGGTGTTGCCAACGAAACAAGACGACTTCTTCCCGTACGCGAGCGACCCGACCGCTTACTGGACCGGGTACTTCACCTCGAGACCGACCACCAAGTACTTCGAGAGAGAGGGGAACGCTTACCTGCAGATGGTGAAACAGCTGCAGGTGTTGTCGAATTTGGAAAGGCACAACGAGTTCATTCTGAACGAACTGAAGAGCGCAATGGGCATAATGCAACACCACGACGCCATCACCGGCACCGAGAAGCAGCACGTCGCTCATGACTACGAAAGGATCCTGAGCACCGCCATCGACGACGCTCTACTCGTCGCCAGACAGGCCTTCAACAAAGCCGCCCAGGGCGACTCCGGGAAGGCTCCGCTGTTCAGCTACGACAGGTGTCACTTCAACGAATCCAGATGCCACACGTCGGAGACCAGCGAAACGTTCATGCGATTAAGAGGATCAGAGTCAGGGTCAGTTGTTGGCCGGATCAGCACCAAGAGGCTCGATGCGGCCTATAAGCCGGCTGTCTTGCAGGTGACCGTTTACAACCCGTTGGCTTGGGGCGTCAGGGAGGCGGTGAAGCTGCCGGTCACGTATGGACACTACGAGGTCTACGCTCCGAATGGCGATAAAATCGTGACGCAACTGGTCGAGATACCGGACGAGGTTAAGCGCATACCGAACCGACGGTCTGAGGCCACTCACGAGTTGGTGTTCATTGCGCACTTACATCCGTTGGGCGTGAAATCGTTCTACGTTAAAAGAGTTTCGACAACGAAAAGGGCAATCGCTAACGGCAGCTATTTGGAATGGATCAGTGGCGAGGGGGTCCGGTCCGATGGCTATTTAGATAGGCTCAATCAGATAGCCGACGTCAAAGACCTCACGAATCTCGGCCTAGATGCTATTGTTGATGAAGGCAAGGAACCTTTGTGGGAAGATGCAAATGGAACTGGTGCGGCTTTCAAAGGGACTATGAAGAAAAGAGAGACGGACGACTTTAAGATATCGTTCGCGTATTACCCGGGCTGCTTGGGGAACAATGAGGTACCCGCTAACCGCAGTTCAGGCGCGTACATCTTCAGGCCGAACTCGACAAAAGCAATCGGTCTGAACGTGGAATCTGTGAGTGAAATTAGCGGCGACCTACTCAGCGAGGTTCGAACTAAGATACGCGGAAACGTTTTGAGCATCAAGCGTGTTTACAAGGAATTCGATTTTATTGAGCACGAGTGGCTCGTGGGCCCGATAAGCGTCGACGACGGCATCGGCAAGGAGTACGTGGTCAAGTACCAGACGGATATAATAAATAATGGAGAGTTTTACACGGATTCAAACGGCCGACAGATGCTGAAGAGGAAACTGAACGCGCGTCCTCAGTGGAACTTGACGTTGACCGAGCCGGTGCCCGGGAATTATTATCCGGTCACGAACGAGATCTACATCGAGAACGGCGAGAGGTTCACGGTCTTGACGGACCGATCGCAAGGCGGAACCTCGCTCGTCGAAGGCGAAGTGGAACTGATGCTGCACAGGAGATTGCTGCACGACGACGCGTTCGGCGTCGGTGAGGCCCTGAACGAAACGGTTCTCGGCTCGGGGCTGGTGGTTCGCGGGAAACACCGGATCCTGCGCTCGAAGGATCCGGTTGAGATCGCGAAAAGCGTTTTCCAAATGCACCTGCCGCCCGTGGTCTTCGTTTCCGACGCCTCAGGCCTCAGCTACGAGCAATGGATGAGGTTGGAGAATTGCAAGAAGTTTGTCAAGACCCCGATGCCTGACGGGCTCCATCTGCTCACCCTGGAGCCTTGGGGCGACCAGATCCTGTTGAGAATCGAAAACTACAAGACCGGTGCCGAAGATTCGACCATCGAAATTGACCTGCAGGGCATATTCAACGGTATCCATATCAAAAGCGTAAAAGAGACAACGCTAGCCGCAAACCAGTGGCTGGACGACTATGATAAGTGGACTTGGAATACGGAAGGAGACTTTTACGAAAGCTTCAACAAGAAATACGGGAGCTTCGAGGCCGTGAACAAAGCTGTCGACGGACCTGACGTCAGAGAGGACGAAGGCCTCAAAATCAAACTTAAAGCGAAGGAGATAAGGTCGTTTATAGCGGATATCGAATATACCGACTAA
Protein
MLVLALLCLLSISSSVPLHDQITPRPGETCGYESCPKTEPGVLNVHIVPHTHDDVGWLKTVDQYYYGSRNNIQKAGVQYILDSVVKELWEDPKRRFIYVETAFFWKWWVRQSRRVKSKVHTLVRQGRLQFVGGAWSMNDEAAVHYQSTIDQFTWGLSKLNETFGYCGLPRVGWQIDPFGHSRELASLLSAMGYDGLFLGRIDYQDKRTRLRDKTMEMVWRGDDDLGKKSDIFTGALYNTYSPPAGFCFDVLCDDEPIVDDAGSPLYNVDNKMTLFLNYVKKQSQHYRTDNVIITMGGDFTYQDAAMWYTNLDKLIEYSNLKAAKDDLKIKLFYSTPNCYLKAIKDANPVLPTKQDDFFPYASDPTAYWTGYFTSRPTTKYFEREGNAYLQMVKQLQVLSNLERHNEFILNELKSAMGIMQHHDAITGTEKQHVAHDYERILSTAIDDALLVARQAFNKAAQGDSGKAPLFSYDRCHFNESRCHTSETSETFMRLRGSESGSVVGRISTKRLDAAYKPAVLQVTVYNPLAWGVREAVKLPVTYGHYEVYAPNGDKIVTQLVEIPDEVKRIPNRRSEATHELVFIAHLHPLGVKSFYVKRVSTTKRAIANGSYLEWISGEGVRSDGYLDRLNQIADVKDLTNLGLDAIVDEGKEPLWEDANGTGAAFKGTMKKRETDDFKISFAYYPGCLGNNEVPANRSSGAYIFRPNSTKAIGLNVESVSEISGDLLSEVRTKIRGNVLSIKRVYKEFDFIEHEWLVGPISVDDGIGKEYVVKYQTDIINNGEFYTDSNGRQMLKRKLNARPQWNLTLTEPVPGNYYPVTNEIYIENGERFTVLTDRSQGGTSLVEGEVELMLHRRLLHDDAFGVGEALNETVLGSGLVVRGKHRILRSKDPVEIAKSVFQMHLPPVVFVSDASGLSYEQWMRLENCKKFVKTPMPDGLHLLTLEPWGDQILLRIENYKTGAEDSTIEIDLQGIFNGIHIKSVKETTLAANQWLDDYDKWTWNTEGDFYESFNKKYGSFEAVNKAVDGPDVREDEGLKIKLKAKEIRSFIADIEYTD
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the glycosyl hydrolase 38 family.
Feature
chain Alpha-mannosidase
Uniprot
A0A2J7RN33
A0A2J7RN15
A0A336LVA7
A0A336LYZ7
A0A1Q3FMY3
U5EV98
+ More
A0A023EX89 A0A1S4FBS2 A0A067R124 A0A1J1I5Z3 A0A336M0Y1 A0A084VFG7 B4JQ04 A0A0C9Q6G3 A0A182IPC8 A0A1Q3FMS6 A0A1J1I212 B3MJI9 A0A2P8Z1T1 A0A1I8M2X1 A0A1S4HD42 Q178V9 A0A182QEX5 A0A0P8XGR3 Q5TS83 A0A084VFG4 A0A182KMH3 A0A182VJW0 Q178W0 A0A1A9ZH71 W5JQI4 A0A1S4GYN8 D6WI18 A0A1W4WS90 A0A3G5BIG9 A0A182IBN6 A0A1A9W6T1 A0A195F0Z8 A0A0K8UEK7 A0A026VXZ2 A0A023EWA7 A0A0K8U3N4 A0A0Q9WJC0 A0A0K8VTI9 A0A182WFK9 A0A1A9XCE3 A0A0Q9XIT5 A0A1I8M2X3 A0A1W4WSD2 A0A182UGZ7 A0A0M4ER29 B4G755 A0A1S4GYP5 A0A1I8QCY1 A0A1B0B0M4 A0A1Q3FJU6 B4Q9B2 A0A1Y1KHF4 A0A0A1WZV2 A0A1W4UG04 Q9VKV1 A0A0J9R086 A0A0K8VT60 E9IED8 A0A182KMH4 A0A0Q5WN53 B4LVB7 A0A1W4UUP0 A0A151IBC4 B3N5F1 Q8IPB7 A0A0R1DR73 A0A182X605 A0A0A1XEF2 A0A2M4DMV2 A0A1I8QD07 A0A195DWW0 A0A034V6I7 W8APD9 J9JL57 A0A0K8WDF9 A0A158NP11 E2B6B9 A0A2H8TPV8 A0A1L8DQQ1 A0A1L8DR39 A0A1W7RA88 A0A2S2R8V4 A0A0P6FNV7 A0A0N8EK80 A0A164XCC5 A0A0P5NKK4 A0A0P5A6R2 A0A0P5Z7T6 A0A0P5ACP0
A0A023EX89 A0A1S4FBS2 A0A067R124 A0A1J1I5Z3 A0A336M0Y1 A0A084VFG7 B4JQ04 A0A0C9Q6G3 A0A182IPC8 A0A1Q3FMS6 A0A1J1I212 B3MJI9 A0A2P8Z1T1 A0A1I8M2X1 A0A1S4HD42 Q178V9 A0A182QEX5 A0A0P8XGR3 Q5TS83 A0A084VFG4 A0A182KMH3 A0A182VJW0 Q178W0 A0A1A9ZH71 W5JQI4 A0A1S4GYN8 D6WI18 A0A1W4WS90 A0A3G5BIG9 A0A182IBN6 A0A1A9W6T1 A0A195F0Z8 A0A0K8UEK7 A0A026VXZ2 A0A023EWA7 A0A0K8U3N4 A0A0Q9WJC0 A0A0K8VTI9 A0A182WFK9 A0A1A9XCE3 A0A0Q9XIT5 A0A1I8M2X3 A0A1W4WSD2 A0A182UGZ7 A0A0M4ER29 B4G755 A0A1S4GYP5 A0A1I8QCY1 A0A1B0B0M4 A0A1Q3FJU6 B4Q9B2 A0A1Y1KHF4 A0A0A1WZV2 A0A1W4UG04 Q9VKV1 A0A0J9R086 A0A0K8VT60 E9IED8 A0A182KMH4 A0A0Q5WN53 B4LVB7 A0A1W4UUP0 A0A151IBC4 B3N5F1 Q8IPB7 A0A0R1DR73 A0A182X605 A0A0A1XEF2 A0A2M4DMV2 A0A1I8QD07 A0A195DWW0 A0A034V6I7 W8APD9 J9JL57 A0A0K8WDF9 A0A158NP11 E2B6B9 A0A2H8TPV8 A0A1L8DQQ1 A0A1L8DR39 A0A1W7RA88 A0A2S2R8V4 A0A0P6FNV7 A0A0N8EK80 A0A164XCC5 A0A0P5NKK4 A0A0P5A6R2 A0A0P5Z7T6 A0A0P5ACP0
EC Number
3.2.1.-
Pubmed
24945155
24845553
24438588
17994087
29403074
25315136
+ More
12364791 17510324 20966253 20920257 23761445 18362917 19820115 30400621 24508170 30249741 22936249 28004739 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 17550304 25348373 24495485 21347285 20798317
12364791 17510324 20966253 20920257 23761445 18362917 19820115 30400621 24508170 30249741 22936249 28004739 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 17550304 25348373 24495485 21347285 20798317
EMBL
NEVH01002545
PNF42229.1
PNF42230.1
UFQS01000218
UFQT01000218
SSX01504.1
+ More
SSX21884.1 SSX01503.1 SSX21883.1 GFDL01006179 JAV28866.1 GANO01001972 JAB57899.1 GAPW01000077 JAC13521.1 KK853110 KDR11229.1 CVRI01000038 CRK93801.1 UFQT01000391 SSX23894.1 ATLV01012390 KE524787 KFB36711.1 CH916372 EDV98984.1 GBYB01009698 JAG79465.1 GFDL01006180 JAV28865.1 CRK93802.1 CH902620 EDV32357.1 PYGN01000238 PSN50451.1 AAAB01008944 CH477358 EAT42740.1 AXCN02000716 KPU73915.1 EAL40242.3 ATLV01012389 KFB36708.1 EAT42739.1 ADMH02000752 ETN65165.1 KQ971332 EEZ99703.1 MK075175 AYV99578.1 APCN01000712 APCN01000713 APCN01000714 APCN01000715 KQ981891 KYN33784.1 GDHF01027225 GDHF01012263 JAI25089.1 JAI40051.1 KK107648 QOIP01000008 EZA48346.1 RLU19783.1 GAPW01000098 JAC13500.1 GDHF01030990 JAI21324.1 CH940649 KRF81022.1 GDHF01010454 JAI41860.1 CH933807 KRG03704.1 CP012523 ALC39282.1 CH479180 EDW28309.1 JXJN01006781 GFDL01007289 JAV27756.1 CM000361 CM002910 EDX04554.1 KMY89555.1 GEZM01085309 JAV60008.1 GBXI01009935 JAD04357.1 AF145601 AE014134 AAD38576.1 AAF52958.2 KMY89556.1 GDHF01010271 GDHF01002021 JAI42043.1 JAI50293.1 GL762576 EFZ21090.1 CH954177 KQS70602.1 EDW63296.2 KQ978112 KYM96921.1 EDV59030.2 AAN10754.1 CM000157 KRJ97497.1 GBXI01004996 JAD09296.1 GGFL01014681 MBW78859.1 KQ980167 KYN17373.1 GAKP01021245 JAC37707.1 GAMC01015917 GAMC01015916 GAMC01015915 GAMC01015914 JAB90640.1 ABLF02026049 GDHF01003182 JAI49132.1 ADTU01022012 ADTU01022013 ADTU01022014 GL445930 EFN88812.1 GFXV01004418 MBW16223.1 GFDF01005288 JAV08796.1 GFDF01005289 JAV08795.1 GFAH01000335 JAV48054.1 GGMS01017236 MBY86439.1 GDIQ01056497 JAN38240.1 GDIQ01018628 JAN76109.1 LRGB01001005 KZS14076.1 GDIQ01144965 JAL06761.1 GDIP01203317 JAJ20085.1 GDIP01060333 JAM43382.1 GDIP01201762 JAJ21640.1
SSX21884.1 SSX01503.1 SSX21883.1 GFDL01006179 JAV28866.1 GANO01001972 JAB57899.1 GAPW01000077 JAC13521.1 KK853110 KDR11229.1 CVRI01000038 CRK93801.1 UFQT01000391 SSX23894.1 ATLV01012390 KE524787 KFB36711.1 CH916372 EDV98984.1 GBYB01009698 JAG79465.1 GFDL01006180 JAV28865.1 CRK93802.1 CH902620 EDV32357.1 PYGN01000238 PSN50451.1 AAAB01008944 CH477358 EAT42740.1 AXCN02000716 KPU73915.1 EAL40242.3 ATLV01012389 KFB36708.1 EAT42739.1 ADMH02000752 ETN65165.1 KQ971332 EEZ99703.1 MK075175 AYV99578.1 APCN01000712 APCN01000713 APCN01000714 APCN01000715 KQ981891 KYN33784.1 GDHF01027225 GDHF01012263 JAI25089.1 JAI40051.1 KK107648 QOIP01000008 EZA48346.1 RLU19783.1 GAPW01000098 JAC13500.1 GDHF01030990 JAI21324.1 CH940649 KRF81022.1 GDHF01010454 JAI41860.1 CH933807 KRG03704.1 CP012523 ALC39282.1 CH479180 EDW28309.1 JXJN01006781 GFDL01007289 JAV27756.1 CM000361 CM002910 EDX04554.1 KMY89555.1 GEZM01085309 JAV60008.1 GBXI01009935 JAD04357.1 AF145601 AE014134 AAD38576.1 AAF52958.2 KMY89556.1 GDHF01010271 GDHF01002021 JAI42043.1 JAI50293.1 GL762576 EFZ21090.1 CH954177 KQS70602.1 EDW63296.2 KQ978112 KYM96921.1 EDV59030.2 AAN10754.1 CM000157 KRJ97497.1 GBXI01004996 JAD09296.1 GGFL01014681 MBW78859.1 KQ980167 KYN17373.1 GAKP01021245 JAC37707.1 GAMC01015917 GAMC01015916 GAMC01015915 GAMC01015914 JAB90640.1 ABLF02026049 GDHF01003182 JAI49132.1 ADTU01022012 ADTU01022013 ADTU01022014 GL445930 EFN88812.1 GFXV01004418 MBW16223.1 GFDF01005288 JAV08796.1 GFDF01005289 JAV08795.1 GFAH01000335 JAV48054.1 GGMS01017236 MBY86439.1 GDIQ01056497 JAN38240.1 GDIQ01018628 JAN76109.1 LRGB01001005 KZS14076.1 GDIQ01144965 JAL06761.1 GDIP01203317 JAJ20085.1 GDIP01060333 JAM43382.1 GDIP01201762 JAJ21640.1
Proteomes
UP000235965
UP000027135
UP000183832
UP000030765
UP000001070
UP000075880
+ More
UP000007801 UP000245037 UP000095301 UP000008820 UP000075886 UP000007062 UP000075882 UP000075903 UP000092445 UP000000673 UP000007266 UP000192223 UP000075840 UP000091820 UP000078541 UP000053097 UP000279307 UP000008792 UP000075920 UP000092443 UP000009192 UP000075902 UP000092553 UP000008744 UP000095300 UP000092460 UP000000304 UP000192221 UP000000803 UP000008711 UP000078542 UP000002282 UP000076407 UP000078492 UP000007819 UP000005205 UP000008237 UP000076858
UP000007801 UP000245037 UP000095301 UP000008820 UP000075886 UP000007062 UP000075882 UP000075903 UP000092445 UP000000673 UP000007266 UP000192223 UP000075840 UP000091820 UP000078541 UP000053097 UP000279307 UP000008792 UP000075920 UP000092443 UP000009192 UP000075902 UP000092553 UP000008744 UP000095300 UP000092460 UP000000304 UP000192221 UP000000803 UP000008711 UP000078542 UP000002282 UP000076407 UP000078492 UP000007819 UP000005205 UP000008237 UP000076858
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A2J7RN33
A0A2J7RN15
A0A336LVA7
A0A336LYZ7
A0A1Q3FMY3
U5EV98
+ More
A0A023EX89 A0A1S4FBS2 A0A067R124 A0A1J1I5Z3 A0A336M0Y1 A0A084VFG7 B4JQ04 A0A0C9Q6G3 A0A182IPC8 A0A1Q3FMS6 A0A1J1I212 B3MJI9 A0A2P8Z1T1 A0A1I8M2X1 A0A1S4HD42 Q178V9 A0A182QEX5 A0A0P8XGR3 Q5TS83 A0A084VFG4 A0A182KMH3 A0A182VJW0 Q178W0 A0A1A9ZH71 W5JQI4 A0A1S4GYN8 D6WI18 A0A1W4WS90 A0A3G5BIG9 A0A182IBN6 A0A1A9W6T1 A0A195F0Z8 A0A0K8UEK7 A0A026VXZ2 A0A023EWA7 A0A0K8U3N4 A0A0Q9WJC0 A0A0K8VTI9 A0A182WFK9 A0A1A9XCE3 A0A0Q9XIT5 A0A1I8M2X3 A0A1W4WSD2 A0A182UGZ7 A0A0M4ER29 B4G755 A0A1S4GYP5 A0A1I8QCY1 A0A1B0B0M4 A0A1Q3FJU6 B4Q9B2 A0A1Y1KHF4 A0A0A1WZV2 A0A1W4UG04 Q9VKV1 A0A0J9R086 A0A0K8VT60 E9IED8 A0A182KMH4 A0A0Q5WN53 B4LVB7 A0A1W4UUP0 A0A151IBC4 B3N5F1 Q8IPB7 A0A0R1DR73 A0A182X605 A0A0A1XEF2 A0A2M4DMV2 A0A1I8QD07 A0A195DWW0 A0A034V6I7 W8APD9 J9JL57 A0A0K8WDF9 A0A158NP11 E2B6B9 A0A2H8TPV8 A0A1L8DQQ1 A0A1L8DR39 A0A1W7RA88 A0A2S2R8V4 A0A0P6FNV7 A0A0N8EK80 A0A164XCC5 A0A0P5NKK4 A0A0P5A6R2 A0A0P5Z7T6 A0A0P5ACP0
A0A023EX89 A0A1S4FBS2 A0A067R124 A0A1J1I5Z3 A0A336M0Y1 A0A084VFG7 B4JQ04 A0A0C9Q6G3 A0A182IPC8 A0A1Q3FMS6 A0A1J1I212 B3MJI9 A0A2P8Z1T1 A0A1I8M2X1 A0A1S4HD42 Q178V9 A0A182QEX5 A0A0P8XGR3 Q5TS83 A0A084VFG4 A0A182KMH3 A0A182VJW0 Q178W0 A0A1A9ZH71 W5JQI4 A0A1S4GYN8 D6WI18 A0A1W4WS90 A0A3G5BIG9 A0A182IBN6 A0A1A9W6T1 A0A195F0Z8 A0A0K8UEK7 A0A026VXZ2 A0A023EWA7 A0A0K8U3N4 A0A0Q9WJC0 A0A0K8VTI9 A0A182WFK9 A0A1A9XCE3 A0A0Q9XIT5 A0A1I8M2X3 A0A1W4WSD2 A0A182UGZ7 A0A0M4ER29 B4G755 A0A1S4GYP5 A0A1I8QCY1 A0A1B0B0M4 A0A1Q3FJU6 B4Q9B2 A0A1Y1KHF4 A0A0A1WZV2 A0A1W4UG04 Q9VKV1 A0A0J9R086 A0A0K8VT60 E9IED8 A0A182KMH4 A0A0Q5WN53 B4LVB7 A0A1W4UUP0 A0A151IBC4 B3N5F1 Q8IPB7 A0A0R1DR73 A0A182X605 A0A0A1XEF2 A0A2M4DMV2 A0A1I8QD07 A0A195DWW0 A0A034V6I7 W8APD9 J9JL57 A0A0K8WDF9 A0A158NP11 E2B6B9 A0A2H8TPV8 A0A1L8DQQ1 A0A1L8DR39 A0A1W7RA88 A0A2S2R8V4 A0A0P6FNV7 A0A0N8EK80 A0A164XCC5 A0A0P5NKK4 A0A0P5A6R2 A0A0P5Z7T6 A0A0P5ACP0
PDB
6B9P
E-value=3.76021e-155,
Score=1410
Ontologies
PATHWAY
GO
Topology
SignalP
Position: 1 - 15,
Likelihood: 0.997810
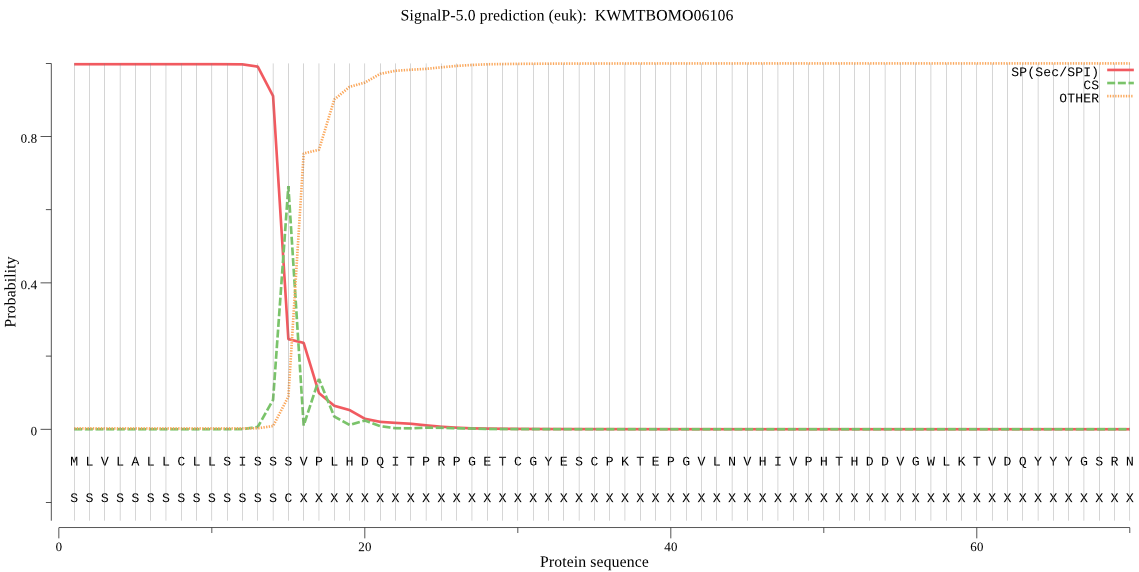
Length:
1057
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0253399999999999
Exp number, first 60 AAs:
0.01927
Total prob of N-in:
0.00104
outside
1 - 1057
Population Genetic Test Statistics
Pi
210.623017
Theta
172.223727
Tajima's D
1.412322
CLR
0.448859
CSRT
0.773311334433278
Interpretation
Uncertain
