Gene
KWMTBOMO06105 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001730
Annotation
PREDICTED:_gelsolin?_cytoplasmic-like_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.735 ER Reliability : 1.341 Nuclear Reliability : 1.084
Sequence
CDS
ATGGAGAATCCGGTGTTTGAGGAGGCAGGAAACCAGGCCGGCATAGAGATCTGGACTATTGAGAACTTCGAGCCGGTTCCCTTTGATCCCAAGTACTATGGGAAATTCTACAACGGCGACTCCTACATCGTGTTGAAGACCACGGAGGAGTCATCCTCGCTGACCCACGACGCCCACTTCTGGCTGGGCAGCAAGACCACTCAGGACAAAAAGGGCTCCGCTGCCATATGGACGATCACGTTAGACGACATGCTGGGCGGCAGAGCTGTGCATCACCGCGAGGTCCAGGGACACGAGTCCAGTTTGTTTCTGAGCTATTTCAAGCCAGCCATCAGATACCTCGAAGGCGGGAACGAGTCTGGCTTCAACGAAGTGCAGACCAACGCTGGTGCTGAGAAACGTCTCCTGAAGCTGTCCGGATGCGACAACATGAGGATCGAGGAGGTCGCAGCCGAGGCCTCGTCTCTGACAAAGAACCACTGCTTCATCTTGGAGAACGAGCACGACATCTTCGTGCTGATGCCGGAGGGCGCGAAAGCCACCCAGAGGCGGAAGATCATCAGCGTCGCCAACAAACTGAGGGACGAGGAACACAACGGGAGAGCCACTATTGAAATCATCGATGAATTCTCGTCAGATGAAGACGTGAATTTGTTCTTCGAAACCCTAGGATCCGGCTCAATGGATGAATTAGTTAGCGACGACACTTCAGAGAGCTACACCAGATCAGATATTTCGGCGGTGTACCTCTACAAAGTGTTGGACGGTGATGAGCAAGAATTAGTCTCCATGAACAAGCCCTACAAGCAGAAACATTTGGCCAGTGACGCAATGTACATTCTGGACACGGGAGACACTGGCGTTTACATCTGGCTGGGGTCGGACCTGGACGCTGACGTTAAGAGGAAATACCACGAGATCGCTGGAAAGTATTTAGAAGACAAGGGATACCCGAGCTGGGTCCACGTGACGAGAGTGTCGGAGGGCGCGGAGAGCTCGACCTTCAAGCAGTACTTCCACAACTGGGACACGGTGCGGACCGGGGGCTCGGTCAGGGGAATCATATCTGAAATCGATTCGGGGTACTTCTCTGGGGACGCTGAGGAGTCGGCGGCCGCGGCCAGGACGATCGGCAAGAGCGCCACAGCACGGGGCTACATGCCGGACCAGGGCGAAGGGAGTCTCACGGTGACCAGGATCTCTGGTGATCAAGAAGACGTGACAGACAGATTCCAGCAGGGGACCTCCGTGCTGTACCAATCAGAGGTCTACGTCATCACTTACAAATACCAGAATGAGAACGAGGAAGATGCCTGGGTGGTTTACTTGTGGATCGGTTCAGAAGCGAGTGATGACGACAAAGAGGCCGGGCAGATGCTACTGTCGCAGCTAGAAGAGGATCTAGAAGGAAATGTTACATCAGTGAAGGTCCCTCAGGGGAAGGAATCGAAGCACTTCTTGAGCATATTCAAAGGAAGCCTCGTGGTGCTTTTTGGTAGCAAAGATGATGATTACCAGCCGCAGAACTCCAAGAAATCGTATGAAGAAGACCAGACGCGGTTGTTCAGGGTGGAGGGAACGGAGACCGGAGTCGACATGAGAGCGGTCCAGGTGGCGGAAGAATCGAACGTTCTAGAAGATGACGACGTATTCGTTCTGGAGACCGGCAGCGGCGTCTATGTTTGGAACGGAAAGGATTCCGACCCTCAAGAACAGGAGGCGGCTCTGAAATTCGTTGAAAATGTAGTCAAGGATAAGGACGTGACGGTGGTGGAACAAGGAGACGAGCCCGAGGAGTTCTGGGAGGCTCTAGGCGGTATACCCGAAGAGAAAGAGGAGCCGTCGGCGTGGCGTATGGTTGTCAACAGACGCGTGACGGCGACACGGACCCTGACAGCTGTCAGTGTCGGCGTCGCCGGGAGGGTTAAGTTCGAGGATCTGGACACGGAATTCTCTCAGCAGGATCTTTCTGAAGATGGAGTTTATATTTTGGACACGGGCGAGGAGCTGTACTTCTGGCAGGGAGCCGACGTCACGGAACGCGTCAAATCCTCCAAACAGGAAGTCATCAAGAAGTACATAGAGGACGACGGTCTCGATCGAACCGTGGACTCGGCGGTGGTCGTGACCGTGAAGCAAGGCGCAGAGCCGCGCGTCTTCAAGAAACTATTTCCCGACTGGGACGACGGAATGTGGAGTAAACTATCATCTTATGAAGACATGAAGAATAGCACTAAAGCTGCAAATTCTAAATAG
Protein
MENPVFEEAGNQAGIEIWTIENFEPVPFDPKYYGKFYNGDSYIVLKTTEESSSLTHDAHFWLGSKTTQDKKGSAAIWTITLDDMLGGRAVHHREVQGHESSLFLSYFKPAIRYLEGGNESGFNEVQTNAGAEKRLLKLSGCDNMRIEEVAAEASSLTKNHCFILENEHDIFVLMPEGAKATQRRKIISVANKLRDEEHNGRATIEIIDEFSSDEDVNLFFETLGSGSMDELVSDDTSESYTRSDISAVYLYKVLDGDEQELVSMNKPYKQKHLASDAMYILDTGDTGVYIWLGSDLDADVKRKYHEIAGKYLEDKGYPSWVHVTRVSEGAESSTFKQYFHNWDTVRTGGSVRGIISEIDSGYFSGDAEESAAAARTIGKSATARGYMPDQGEGSLTVTRISGDQEDVTDRFQQGTSVLYQSEVYVITYKYQNENEEDAWVVYLWIGSEASDDDKEAGQMLLSQLEEDLEGNVTSVKVPQGKESKHFLSIFKGSLVVLFGSKDDDYQPQNSKKSYEEDQTRLFRVEGTETGVDMRAVQVAEESNVLEDDDVFVLETGSGVYVWNGKDSDPQEQEAALKFVENVVKDKDVTVVEQGDEPEEFWEALGGIPEEKEEPSAWRMVVNRRVTATRTLTAVSVGVAGRVKFEDLDTEFSQQDLSEDGVYILDTGEELYFWQGADVTERVKSSKQEVIKKYIEDDGLDRTVDSAVVVTVKQGAEPRVFKKLFPDWDDGMWSKLSSYEDMKNSTKAANSK
Summary
Uniprot
H9IWU7
A0A2W1BHR2
A0A1E1W2G3
A0A2H1VYX6
A0A1E1WAQ0
A0A2A4K3R4
+ More
A0A212EZ89 B0WBT0 A0A1Q3FQ94 U5ETK7 A0A1Q3FQS9 A0A2J7RA50 A0A1Q3FQD7 A0A1Q3FQB1 A0A1Q3FQM8 A0A2J7RA31 A0A1S4FCM2 Q177K8 Q177L5 A0A336MRC3 A0A1S4FCS4 Q177K7 Q177L4 A0A336MY83 A0A1J1HKI1 A0A084WAH4 A0A1L8E5M0 A0A336MF08 A0A1L8E606 A0A1L8E5D0 A0A2J7RA37 A0A1L8E5G4 A0A182JFK4 A0A1L8E5N5 A0A1L8E5B4 W5JRC5 A0A0P4W1A2 A0A0P5HP29 A0A1S4HE98 A0A182QRY7 A0A0N8BCY3 A0A0P4W7B4 B4KCW4 I7L125 A0A0Q9XDT7 A0A0P5EIR4 B4NFJ3 A0A0P5CZ52 A0A0P5EMU7 A0A182HY18 A0A034VMT8 A0A182PGI3 T1IZM9 B4JS24 A0A182WXE2 Q5TPW1 A0A182FPK8 A0A182RVF1 I7ITS8 E9FTN2 A0A0P6BTR4 A0A1Q3FPV9 A0A0M3QXF2 A0A182W1Q4 A0A336M909 A0A0K8VEK4 I5APK8 A0A1Q3FQ87 B4GM71 Q295D4 A0A0Q9XFV1 A0A0P6F3G8 W8BFB5 A0A0P6GG72 A0A0A1XCB7 A0A034VNT3 A0A0P5GHB5 A0A224YQ72 A0A182TJF0 A0A0Q9WBU6 B4MBI4 A0A0P5SCV8 A0A0P4ZK42 A0A182K0I4 A0A1W4VZL6 A0A1W4VLN5 A0A0P5K9Q4 A0A182Y734 A0A1I8NXE8 A0A1I8NXC0 A0A2M4CWF0 W8B6S7 A0A0K8WH15 A0A0N8P1H5 A0A1E1XAG7 A0A0T6ATX5 A0A182VE77 A0A293LI79 B3LXY4
A0A212EZ89 B0WBT0 A0A1Q3FQ94 U5ETK7 A0A1Q3FQS9 A0A2J7RA50 A0A1Q3FQD7 A0A1Q3FQB1 A0A1Q3FQM8 A0A2J7RA31 A0A1S4FCM2 Q177K8 Q177L5 A0A336MRC3 A0A1S4FCS4 Q177K7 Q177L4 A0A336MY83 A0A1J1HKI1 A0A084WAH4 A0A1L8E5M0 A0A336MF08 A0A1L8E606 A0A1L8E5D0 A0A2J7RA37 A0A1L8E5G4 A0A182JFK4 A0A1L8E5N5 A0A1L8E5B4 W5JRC5 A0A0P4W1A2 A0A0P5HP29 A0A1S4HE98 A0A182QRY7 A0A0N8BCY3 A0A0P4W7B4 B4KCW4 I7L125 A0A0Q9XDT7 A0A0P5EIR4 B4NFJ3 A0A0P5CZ52 A0A0P5EMU7 A0A182HY18 A0A034VMT8 A0A182PGI3 T1IZM9 B4JS24 A0A182WXE2 Q5TPW1 A0A182FPK8 A0A182RVF1 I7ITS8 E9FTN2 A0A0P6BTR4 A0A1Q3FPV9 A0A0M3QXF2 A0A182W1Q4 A0A336M909 A0A0K8VEK4 I5APK8 A0A1Q3FQ87 B4GM71 Q295D4 A0A0Q9XFV1 A0A0P6F3G8 W8BFB5 A0A0P6GG72 A0A0A1XCB7 A0A034VNT3 A0A0P5GHB5 A0A224YQ72 A0A182TJF0 A0A0Q9WBU6 B4MBI4 A0A0P5SCV8 A0A0P4ZK42 A0A182K0I4 A0A1W4VZL6 A0A1W4VLN5 A0A0P5K9Q4 A0A182Y734 A0A1I8NXE8 A0A1I8NXC0 A0A2M4CWF0 W8B6S7 A0A0K8WH15 A0A0N8P1H5 A0A1E1XAG7 A0A0T6ATX5 A0A182VE77 A0A293LI79 B3LXY4
Pubmed
EMBL
BABH01020975
KZ150158
PZC72747.1
GDQN01009920
JAT81134.1
ODYU01004997
+ More
SOQ45434.1 GDQN01006982 JAT84072.1 NWSH01000217 PCG78292.1 AGBW02011330 OWR46816.1 DS231882 EDS42860.1 GFDL01005264 JAV29781.1 GANO01002735 JAB57136.1 GFDL01005187 JAV29858.1 NEVH01006578 PNF37694.1 GFDL01005226 JAV29819.1 GFDL01005244 JAV29801.1 GFDL01005237 JAV29808.1 PNF37691.1 CH477374 EAT42359.1 EAT42351.1 UFQT01001498 SSX30817.1 EAT42358.1 EAT42352.1 SSX30818.1 CVRI01000008 CRK88567.1 ATLV01022193 KE525329 KFB47218.1 GFDF01000140 JAV13944.1 UFQS01001020 UFQT01001020 SSX08604.1 SSX28520.1 GFDF01000142 JAV13942.1 GFDF01000143 JAV13941.1 PNF37692.1 GFDF01000141 JAV13943.1 GFDF01000145 JAV13939.1 GFDF01000144 JAV13940.1 ADMH02000569 ETN65858.1 GDRN01080643 JAI62170.1 GDIQ01226848 JAK24877.1 AAAB01008966 AXCN02001122 GDIQ01189995 JAK61730.1 GDRN01080645 JAI62169.1 CH933806 EDW16986.1 KRG02434.1 HE861499 CCI71879.1 KRG02433.1 GDIQ01271127 JAJ80597.1 CH964251 EDW83060.2 GDIP01163430 JAJ59972.1 GDIP01157436 JAJ65966.1 GAKP01015189 GAKP01015188 JAC43764.1 JH431720 CH916373 EDV94564.1 EAL39527.3 HE861500 CCI71880.1 GL732524 EFX89391.1 GDIP01011124 JAM92591.1 GFDL01005406 JAV29639.1 CP012526 ALC45823.1 UFQT01000511 SSX24857.1 GDHF01015067 GDHF01010278 JAI37247.1 JAI42036.1 CM000070 EIM52893.1 GFDL01005341 JAV29704.1 CH479185 EDW37945.1 EAL28778.1 KRG02435.1 GDIQ01068363 JAN26374.1 GAMC01006570 JAB99985.1 GDIQ01035411 JAN59326.1 GBXI01005705 JAD08587.1 GAKP01015191 GAKP01015190 JAC43762.1 GDIQ01240786 JAK10939.1 GFPF01007949 MAA19095.1 CH940656 KRF78299.1 EDW58455.1 GDIP01141281 JAL62433.1 GDIP01212226 JAJ11176.1 GDIQ01187255 JAK64470.1 GGFL01005472 MBW69650.1 GAMC01012223 JAB94332.1 GDHF01032030 GDHF01001868 JAI20284.1 JAI50446.1 CH902617 KPU79940.1 GFAC01002976 JAT96212.1 LJIG01022826 KRT78520.1 GFWV01008468 MAA33197.1 EDV42840.1
SOQ45434.1 GDQN01006982 JAT84072.1 NWSH01000217 PCG78292.1 AGBW02011330 OWR46816.1 DS231882 EDS42860.1 GFDL01005264 JAV29781.1 GANO01002735 JAB57136.1 GFDL01005187 JAV29858.1 NEVH01006578 PNF37694.1 GFDL01005226 JAV29819.1 GFDL01005244 JAV29801.1 GFDL01005237 JAV29808.1 PNF37691.1 CH477374 EAT42359.1 EAT42351.1 UFQT01001498 SSX30817.1 EAT42358.1 EAT42352.1 SSX30818.1 CVRI01000008 CRK88567.1 ATLV01022193 KE525329 KFB47218.1 GFDF01000140 JAV13944.1 UFQS01001020 UFQT01001020 SSX08604.1 SSX28520.1 GFDF01000142 JAV13942.1 GFDF01000143 JAV13941.1 PNF37692.1 GFDF01000141 JAV13943.1 GFDF01000145 JAV13939.1 GFDF01000144 JAV13940.1 ADMH02000569 ETN65858.1 GDRN01080643 JAI62170.1 GDIQ01226848 JAK24877.1 AAAB01008966 AXCN02001122 GDIQ01189995 JAK61730.1 GDRN01080645 JAI62169.1 CH933806 EDW16986.1 KRG02434.1 HE861499 CCI71879.1 KRG02433.1 GDIQ01271127 JAJ80597.1 CH964251 EDW83060.2 GDIP01163430 JAJ59972.1 GDIP01157436 JAJ65966.1 GAKP01015189 GAKP01015188 JAC43764.1 JH431720 CH916373 EDV94564.1 EAL39527.3 HE861500 CCI71880.1 GL732524 EFX89391.1 GDIP01011124 JAM92591.1 GFDL01005406 JAV29639.1 CP012526 ALC45823.1 UFQT01000511 SSX24857.1 GDHF01015067 GDHF01010278 JAI37247.1 JAI42036.1 CM000070 EIM52893.1 GFDL01005341 JAV29704.1 CH479185 EDW37945.1 EAL28778.1 KRG02435.1 GDIQ01068363 JAN26374.1 GAMC01006570 JAB99985.1 GDIQ01035411 JAN59326.1 GBXI01005705 JAD08587.1 GAKP01015191 GAKP01015190 JAC43762.1 GDIQ01240786 JAK10939.1 GFPF01007949 MAA19095.1 CH940656 KRF78299.1 EDW58455.1 GDIP01141281 JAL62433.1 GDIP01212226 JAJ11176.1 GDIQ01187255 JAK64470.1 GGFL01005472 MBW69650.1 GAMC01012223 JAB94332.1 GDHF01032030 GDHF01001868 JAI20284.1 JAI50446.1 CH902617 KPU79940.1 GFAC01002976 JAT96212.1 LJIG01022826 KRT78520.1 GFWV01008468 MAA33197.1 EDV42840.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000002320
UP000235965
UP000008820
+ More
UP000183832 UP000030765 UP000075880 UP000000673 UP000075886 UP000009192 UP000007798 UP000075885 UP000001070 UP000076407 UP000007062 UP000069272 UP000075900 UP000000305 UP000092553 UP000075920 UP000001819 UP000008744 UP000075902 UP000008792 UP000075881 UP000192221 UP000076408 UP000095300 UP000007801 UP000075903
UP000183832 UP000030765 UP000075880 UP000000673 UP000075886 UP000009192 UP000007798 UP000075885 UP000001070 UP000076407 UP000007062 UP000069272 UP000075900 UP000000305 UP000092553 UP000075920 UP000001819 UP000008744 UP000075902 UP000008792 UP000075881 UP000192221 UP000076408 UP000095300 UP000007801 UP000075903
PRIDE
Pfam
PF00626 Gelsolin
Interpro
SUPFAM
SSF82754
SSF82754
Gene 3D
ProteinModelPortal
H9IWU7
A0A2W1BHR2
A0A1E1W2G3
A0A2H1VYX6
A0A1E1WAQ0
A0A2A4K3R4
+ More
A0A212EZ89 B0WBT0 A0A1Q3FQ94 U5ETK7 A0A1Q3FQS9 A0A2J7RA50 A0A1Q3FQD7 A0A1Q3FQB1 A0A1Q3FQM8 A0A2J7RA31 A0A1S4FCM2 Q177K8 Q177L5 A0A336MRC3 A0A1S4FCS4 Q177K7 Q177L4 A0A336MY83 A0A1J1HKI1 A0A084WAH4 A0A1L8E5M0 A0A336MF08 A0A1L8E606 A0A1L8E5D0 A0A2J7RA37 A0A1L8E5G4 A0A182JFK4 A0A1L8E5N5 A0A1L8E5B4 W5JRC5 A0A0P4W1A2 A0A0P5HP29 A0A1S4HE98 A0A182QRY7 A0A0N8BCY3 A0A0P4W7B4 B4KCW4 I7L125 A0A0Q9XDT7 A0A0P5EIR4 B4NFJ3 A0A0P5CZ52 A0A0P5EMU7 A0A182HY18 A0A034VMT8 A0A182PGI3 T1IZM9 B4JS24 A0A182WXE2 Q5TPW1 A0A182FPK8 A0A182RVF1 I7ITS8 E9FTN2 A0A0P6BTR4 A0A1Q3FPV9 A0A0M3QXF2 A0A182W1Q4 A0A336M909 A0A0K8VEK4 I5APK8 A0A1Q3FQ87 B4GM71 Q295D4 A0A0Q9XFV1 A0A0P6F3G8 W8BFB5 A0A0P6GG72 A0A0A1XCB7 A0A034VNT3 A0A0P5GHB5 A0A224YQ72 A0A182TJF0 A0A0Q9WBU6 B4MBI4 A0A0P5SCV8 A0A0P4ZK42 A0A182K0I4 A0A1W4VZL6 A0A1W4VLN5 A0A0P5K9Q4 A0A182Y734 A0A1I8NXE8 A0A1I8NXC0 A0A2M4CWF0 W8B6S7 A0A0K8WH15 A0A0N8P1H5 A0A1E1XAG7 A0A0T6ATX5 A0A182VE77 A0A293LI79 B3LXY4
A0A212EZ89 B0WBT0 A0A1Q3FQ94 U5ETK7 A0A1Q3FQS9 A0A2J7RA50 A0A1Q3FQD7 A0A1Q3FQB1 A0A1Q3FQM8 A0A2J7RA31 A0A1S4FCM2 Q177K8 Q177L5 A0A336MRC3 A0A1S4FCS4 Q177K7 Q177L4 A0A336MY83 A0A1J1HKI1 A0A084WAH4 A0A1L8E5M0 A0A336MF08 A0A1L8E606 A0A1L8E5D0 A0A2J7RA37 A0A1L8E5G4 A0A182JFK4 A0A1L8E5N5 A0A1L8E5B4 W5JRC5 A0A0P4W1A2 A0A0P5HP29 A0A1S4HE98 A0A182QRY7 A0A0N8BCY3 A0A0P4W7B4 B4KCW4 I7L125 A0A0Q9XDT7 A0A0P5EIR4 B4NFJ3 A0A0P5CZ52 A0A0P5EMU7 A0A182HY18 A0A034VMT8 A0A182PGI3 T1IZM9 B4JS24 A0A182WXE2 Q5TPW1 A0A182FPK8 A0A182RVF1 I7ITS8 E9FTN2 A0A0P6BTR4 A0A1Q3FPV9 A0A0M3QXF2 A0A182W1Q4 A0A336M909 A0A0K8VEK4 I5APK8 A0A1Q3FQ87 B4GM71 Q295D4 A0A0Q9XFV1 A0A0P6F3G8 W8BFB5 A0A0P6GG72 A0A0A1XCB7 A0A034VNT3 A0A0P5GHB5 A0A224YQ72 A0A182TJF0 A0A0Q9WBU6 B4MBI4 A0A0P5SCV8 A0A0P4ZK42 A0A182K0I4 A0A1W4VZL6 A0A1W4VLN5 A0A0P5K9Q4 A0A182Y734 A0A1I8NXE8 A0A1I8NXC0 A0A2M4CWF0 W8B6S7 A0A0K8WH15 A0A0N8P1H5 A0A1E1XAG7 A0A0T6ATX5 A0A182VE77 A0A293LI79 B3LXY4
PDB
2FGH
E-value=2.2757e-100,
Score=936
Ontologies
GO
PANTHER
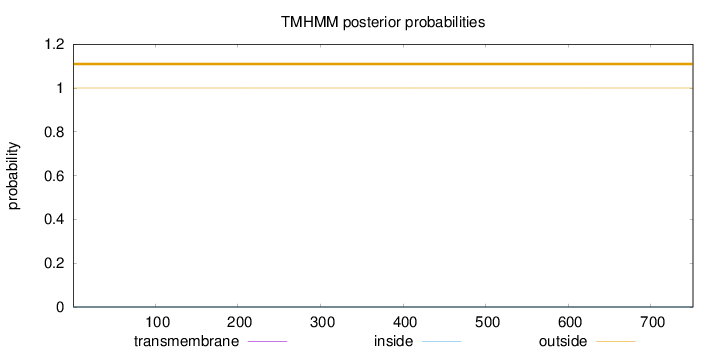
Topology
Length:
751
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00111
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00004
outside
1 - 751
Population Genetic Test Statistics
Pi
18.61216
Theta
154.495009
Tajima's D
0.017031
CLR
0.405899
CSRT
0.373881305934703
Interpretation
Uncertain
