Gene
KWMTBOMO06102
Pre Gene Modal
BGIBMGA001698
Annotation
PREDICTED:_copper_chaperone_for_superoxide_dismutase_[Papilio_machaon]
Full name
Superoxide dismutase [Cu-Zn]
+ More
Copper chaperone for superoxide dismutase
Copper chaperone for superoxide dismutase
Alternative Name
Superoxide dismutase copper chaperone
Location in the cell
Cytoplasmic Reliability : 1.894 Extracellular Reliability : 1.579
Sequence
CDS
ATGCTGACTACTAAGCTAGAGGTCCTGGTGAACTTCGATTCATCTGTTGATGACCAGTTCCTCGAGAAGACTATTAAGACATTGAAAACTCACGACGGAATCAAGGAGGTGGGTTTCAAGGACGGTGCTTTTGTCGTGGAGACGGTGCTCCCAAGCGCGAACGTTCTGGACGTGGTCGGCTCCGTGACGGGGAGACCAGCCGTGATCCAAGGTTTTGGGGAAACACAATCGGCCGTCGCCATGGTCTCGGACCAATCGTGCTGCAATAAGATCCTGGGCGTGGTCAGGCTGCAGCAGACGGCCGAGGGCCCGCTGGTGGCTGACGGCAGCATCGACGGACTAAGCCCCGGGAGGCACGGGCTCCACGTATATGAGTCCGGGGATTTAAGTCAGGGCTGCAATTCCATAGGGGGGCATTATAATCCGTCGGGGGGACCGCACGGGGGGCCCGGCGATCCCCCGGATCGGAGGCACGCCGGCGATCTCGGAAACATTACAGCCGACGCAAACGGCAGAGCTGCATTCAGGATTGTCGATGATGTCCTAAAAGTGTGGGACGTGATCGGTAGGTCTATGGGCGTGACCGAGCGGGGGGACGACTGCGGCCGCGGAGACGGCACCTCCAGGGTCGACGGGAACTCGGGACCGATATTAGCGTGTGGCATCATAGCCAGGTCGGCGGGAATCTTTCAGAATCCGAAGCGGATATGCGCCTGCGACGGGGTAGTGGTGTGGGATGAACGAGATAAACCGCTGGCGGGCAGCGGGAGGAGAGGTCAGGGGGACCGCGCCCCCAGCGAGGGGGGCTGCTGTGGCAAGGCAGGCAAAACCACGAAAGGAAGTAATAACAAAGGGGACCGATGTGGGGCAGGGGACGGTGGGGGTGGCAACGGTAAAACATGCTGTAAAGTGTAG
Protein
MLTTKLEVLVNFDSSVDDQFLEKTIKTLKTHDGIKEVGFKDGAFVVETVLPSANVLDVVGSVTGRPAVIQGFGETQSAVAMVSDQSCCNKILGVVRLQQTAEGPLVADGSIDGLSPGRHGLHVYESGDLSQGCNSIGGHYNPSGGPHGGPGDPPDRRHAGDLGNITADANGRAAFRIVDDVLKVWDVIGRSMGVTERGDDCGRGDGTSRVDGNSGPILACGIIARSAGIFQNPKRICACDGVVVWDERDKPLAGSGRRGQGDRAPSEGGCCGKAGKTTKGSNNKGDRCGAGDGGGGNGKTCCKV
Summary
Description
Destroys radicals which are normally produced within the cells and which are toxic to biological systems.
Delivers copper to copper zinc superoxide dismutase (SOD1).
Delivers copper to copper zinc superoxide dismutase (SOD1).
Catalytic Activity
2 H(+) + 2 superoxide = H2O2 + O2
Cofactor
Cu cation
Zn(2+)
Cu(2+)
Zn(2+)
Cu(2+)
Subunit
Homodimer, and heterodimer with SOD1. Interacts with COMMD1 (By similarity). Interacts with XIAP/BIRC4 (By similarity).
Similarity
Belongs to the Cu-Zn superoxide dismutase family.
In the C-terminal section; belongs to the Cu-Zn superoxide dismutase family.
In the C-terminal section; belongs to the Cu-Zn superoxide dismutase family.
Keywords
Chaperone
Complete proteome
Copper
Cytoplasm
Disulfide bond
Isopeptide bond
Metal-binding
Phosphoprotein
Reference proteome
Ubl conjugation
Zinc
Feature
chain Copper chaperone for superoxide dismutase
Uniprot
H9IWR6
A0A2A4IVF1
A0A2H1V0V1
A0A0N1I5U1
A0A0N1IG53
S4P4K3
+ More
A0A212ENZ9 A0A1B6G5Y1 A0A088A6F0 A0A2A3E370 A0A0L7R8J2 A0A158P3W4 A0A0M8ZPP8 A0A224XLJ0 A0A069DQU5 K7IYR3 C3Z3X0 A0A023F796 A0A067R164 E2BK27 A0A2J7R1W1 A0A151J8W2 R4G7S6 A0A0J7N4P1 A0A195CNY2 F4WKV5 E2AUD2 A0A2H8TIP7 A0A151WVL5 A0A195BKR7 A0A1U7Q4Z1 J9JTJ9 G3I4K4 A0A1A9YE99 A0A1A9W9Q9 A0A151JVJ1 A0A3L8DQH2 A0A154PQE2 A0A0A9WBK0 A0A026W1C0 A0A1B0AZL2 A0A1A6HW90 Q543K2 Q9WU84 A0A060YNP3 A0A0P4VXF6 A0A1U7T321 A0A0G2JSZ9 I3ME75 A0A3L7HWE1 Q9JK72 A0A0R4IHZ8 A0A0F8AKW0 F1QCS3 A0A2K8FTM4 A0A1A9VID8 A0A2K6F7Q4 D3TN65 W8C4D0 F6YJP8 L8Y595 A0A061IBV5 A0A2Y9JU61 A0A3Q2FVL3 A0A3B4DU04 A0A2K6T6L0 A0A1A7YS36 A0A1A7WRV0 A0A250YE74 B4LJV9 A0A2U9B0H9 A0A091DBB1 A0A0Q9W625 B5X3L5 A0A3Q3K2N1 A0A1A8GPI5 A0A2Y9JZY3 A0A1A8FIV1 A0A2K5RIY8 A0A1A8BXI3 A0A1A8UMW4 A0A1L8GK44 A0A1A7ZTS6 A0A1A8KT13 A0A1A8I878 A0A3G1PVI3 A0A2U3YVZ7 A0A2J8QBB0 B3MHH4 H0WLC7 A0A1A8QGN1 A0A1A8L3W1 J3KNF4 A0A1A8SF48 H2Q473 G3R2G0 A0A286ZIZ7 A0A2J8U030
A0A212ENZ9 A0A1B6G5Y1 A0A088A6F0 A0A2A3E370 A0A0L7R8J2 A0A158P3W4 A0A0M8ZPP8 A0A224XLJ0 A0A069DQU5 K7IYR3 C3Z3X0 A0A023F796 A0A067R164 E2BK27 A0A2J7R1W1 A0A151J8W2 R4G7S6 A0A0J7N4P1 A0A195CNY2 F4WKV5 E2AUD2 A0A2H8TIP7 A0A151WVL5 A0A195BKR7 A0A1U7Q4Z1 J9JTJ9 G3I4K4 A0A1A9YE99 A0A1A9W9Q9 A0A151JVJ1 A0A3L8DQH2 A0A154PQE2 A0A0A9WBK0 A0A026W1C0 A0A1B0AZL2 A0A1A6HW90 Q543K2 Q9WU84 A0A060YNP3 A0A0P4VXF6 A0A1U7T321 A0A0G2JSZ9 I3ME75 A0A3L7HWE1 Q9JK72 A0A0R4IHZ8 A0A0F8AKW0 F1QCS3 A0A2K8FTM4 A0A1A9VID8 A0A2K6F7Q4 D3TN65 W8C4D0 F6YJP8 L8Y595 A0A061IBV5 A0A2Y9JU61 A0A3Q2FVL3 A0A3B4DU04 A0A2K6T6L0 A0A1A7YS36 A0A1A7WRV0 A0A250YE74 B4LJV9 A0A2U9B0H9 A0A091DBB1 A0A0Q9W625 B5X3L5 A0A3Q3K2N1 A0A1A8GPI5 A0A2Y9JZY3 A0A1A8FIV1 A0A2K5RIY8 A0A1A8BXI3 A0A1A8UMW4 A0A1L8GK44 A0A1A7ZTS6 A0A1A8KT13 A0A1A8I878 A0A3G1PVI3 A0A2U3YVZ7 A0A2J8QBB0 B3MHH4 H0WLC7 A0A1A8QGN1 A0A1A8L3W1 J3KNF4 A0A1A8SF48 H2Q473 G3R2G0 A0A286ZIZ7 A0A2J8U030
EC Number
1.15.1.1
Pubmed
19121390
26354079
23622113
22118469
21347285
26334808
+ More
20075255 18563158 25474469 24845553 20798317 21719571 21804562 30249741 25401762 26823975 24508170 10349636 11042159 11076861 11217851 12466851 12040188 16141073 21183079 10694572 10773661 15489334 16141072 24755649 27129103 15057822 15632090 29704459 10964676 23594743 25835551 20353571 24495485 17495919 23385571 23929341 28087693 17994087 20433749 27762356 28965584 16554811 21269460 24275569 16136131 22398555
20075255 18563158 25474469 24845553 20798317 21719571 21804562 30249741 25401762 26823975 24508170 10349636 11042159 11076861 11217851 12466851 12040188 16141073 21183079 10694572 10773661 15489334 16141072 24755649 27129103 15057822 15632090 29704459 10964676 23594743 25835551 20353571 24495485 17495919 23385571 23929341 28087693 17994087 20433749 27762356 28965584 16554811 21269460 24275569 16136131 22398555
EMBL
BABH01020973
BABH01020974
NWSH01007082
PCG63103.1
ODYU01000153
SOQ34441.1
+ More
KQ458725 KPJ05369.1 KQ460546 KPJ14045.1 GAIX01005929 JAA86631.1 AGBW02013590 OWR43196.1 GECZ01011968 JAS57801.1 KZ288405 PBC26155.1 KQ414632 KOC67190.1 ADTU01008606 KQ435922 KOX68555.1 GFTR01004472 JAW11954.1 GBGD01002466 JAC86423.1 GG666577 EEN52884.1 GBBI01001597 JAC17115.1 KK852818 KDR15674.1 GL448740 EFN83935.1 NEVH01008202 PNF34824.1 KQ979480 KYN21259.1 GAHY01002207 JAA75303.1 LBMM01010188 KMQ87620.1 KQ977513 KYN02187.1 GL888206 EGI65197.1 GL442815 EFN62989.1 GFXV01002202 MBW14007.1 KQ982708 KYQ51807.1 KQ976453 KYM85320.1 ABLF02025275 JH001255 EGW06367.1 KQ981701 KYN37474.1 QOIP01000005 RLU22631.1 KQ434998 KZC13330.1 GBHO01038485 GBRD01011502 GDHC01008675 JAG05119.1 JAG54322.1 JAQ09954.1 KK107570 EZA48859.1 JXJN01006362 JXJN01006363 LZPO01008032 OBS82506.1 AK050072 CH466612 BAC34057.1 EDL33071.1 AF121906 AF173379 AK010264 BC026938 FR914611 CDQ93182.1 GDKW01002328 JAI54267.1 AC126581 CH473953 EDM12426.1 AGTP01072334 RAZU01000139 RLQ70153.1 AF255305 BC158586 FO904860 KQ041578 KKF24683.1 BX511071 KY618831 AQW43017.1 EZ422867 ADD19143.1 GAMC01002377 JAC04179.1 KB370292 ELV09526.1 KE673007 ERE78744.1 HADX01011145 SBP33377.1 HADW01006981 SBP08381.1 GFFW01002857 JAV41931.1 CH940648 EDW61613.1 CP026244 AWO97434.1 KN122776 KFO28362.1 KRF80102.1 BT045634 ACI33896.1 HAEC01004844 SBQ72921.1 HAEB01012833 SBQ59360.1 HADZ01007274 SBP71215.1 HAEJ01008205 SBS48662.1 CM004472 OCT84183.1 HADY01006875 SBP45360.1 HAEE01015487 SBR35537.1 HAED01007070 SBQ93251.1 KY660271 AVQ09312.1 NBAG03000050 PNI93550.1 CH902619 EDV37974.1 AAQR03144760 AAQR03144761 HAEG01012692 SBR93005.1 HAEF01002108 SBR39490.1 AP001157 HAEI01013815 SBS16284.1 AACZ04016225 GABC01004358 JAA06980.1 CABD030079815 AEMK02000006 NDHI03003478 PNJ38629.1
KQ458725 KPJ05369.1 KQ460546 KPJ14045.1 GAIX01005929 JAA86631.1 AGBW02013590 OWR43196.1 GECZ01011968 JAS57801.1 KZ288405 PBC26155.1 KQ414632 KOC67190.1 ADTU01008606 KQ435922 KOX68555.1 GFTR01004472 JAW11954.1 GBGD01002466 JAC86423.1 GG666577 EEN52884.1 GBBI01001597 JAC17115.1 KK852818 KDR15674.1 GL448740 EFN83935.1 NEVH01008202 PNF34824.1 KQ979480 KYN21259.1 GAHY01002207 JAA75303.1 LBMM01010188 KMQ87620.1 KQ977513 KYN02187.1 GL888206 EGI65197.1 GL442815 EFN62989.1 GFXV01002202 MBW14007.1 KQ982708 KYQ51807.1 KQ976453 KYM85320.1 ABLF02025275 JH001255 EGW06367.1 KQ981701 KYN37474.1 QOIP01000005 RLU22631.1 KQ434998 KZC13330.1 GBHO01038485 GBRD01011502 GDHC01008675 JAG05119.1 JAG54322.1 JAQ09954.1 KK107570 EZA48859.1 JXJN01006362 JXJN01006363 LZPO01008032 OBS82506.1 AK050072 CH466612 BAC34057.1 EDL33071.1 AF121906 AF173379 AK010264 BC026938 FR914611 CDQ93182.1 GDKW01002328 JAI54267.1 AC126581 CH473953 EDM12426.1 AGTP01072334 RAZU01000139 RLQ70153.1 AF255305 BC158586 FO904860 KQ041578 KKF24683.1 BX511071 KY618831 AQW43017.1 EZ422867 ADD19143.1 GAMC01002377 JAC04179.1 KB370292 ELV09526.1 KE673007 ERE78744.1 HADX01011145 SBP33377.1 HADW01006981 SBP08381.1 GFFW01002857 JAV41931.1 CH940648 EDW61613.1 CP026244 AWO97434.1 KN122776 KFO28362.1 KRF80102.1 BT045634 ACI33896.1 HAEC01004844 SBQ72921.1 HAEB01012833 SBQ59360.1 HADZ01007274 SBP71215.1 HAEJ01008205 SBS48662.1 CM004472 OCT84183.1 HADY01006875 SBP45360.1 HAEE01015487 SBR35537.1 HAED01007070 SBQ93251.1 KY660271 AVQ09312.1 NBAG03000050 PNI93550.1 CH902619 EDV37974.1 AAQR03144760 AAQR03144761 HAEG01012692 SBR93005.1 HAEF01002108 SBR39490.1 AP001157 HAEI01013815 SBS16284.1 AACZ04016225 GABC01004358 JAA06980.1 CABD030079815 AEMK02000006 NDHI03003478 PNJ38629.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000005203
+ More
UP000242457 UP000053825 UP000005205 UP000053105 UP000002358 UP000001554 UP000027135 UP000008237 UP000235965 UP000078492 UP000036403 UP000078542 UP000007755 UP000000311 UP000075809 UP000078540 UP000189706 UP000007819 UP000001075 UP000092443 UP000091820 UP000078541 UP000279307 UP000076502 UP000053097 UP000092460 UP000092124 UP000000589 UP000193380 UP000189704 UP000002494 UP000005215 UP000273346 UP000000437 UP000078200 UP000233160 UP000002280 UP000011518 UP000030759 UP000248482 UP000265020 UP000261440 UP000233220 UP000008792 UP000246464 UP000028990 UP000087266 UP000261600 UP000233040 UP000186698 UP000245341 UP000007801 UP000005225 UP000005640 UP000002277 UP000001519 UP000008227
UP000242457 UP000053825 UP000005205 UP000053105 UP000002358 UP000001554 UP000027135 UP000008237 UP000235965 UP000078492 UP000036403 UP000078542 UP000007755 UP000000311 UP000075809 UP000078540 UP000189706 UP000007819 UP000001075 UP000092443 UP000091820 UP000078541 UP000279307 UP000076502 UP000053097 UP000092460 UP000092124 UP000000589 UP000193380 UP000189704 UP000002494 UP000005215 UP000273346 UP000000437 UP000078200 UP000233160 UP000002280 UP000011518 UP000030759 UP000248482 UP000265020 UP000261440 UP000233220 UP000008792 UP000246464 UP000028990 UP000087266 UP000261600 UP000233040 UP000186698 UP000245341 UP000007801 UP000005225 UP000005640 UP000002277 UP000001519 UP000008227
Interpro
Gene 3D
ProteinModelPortal
H9IWR6
A0A2A4IVF1
A0A2H1V0V1
A0A0N1I5U1
A0A0N1IG53
S4P4K3
+ More
A0A212ENZ9 A0A1B6G5Y1 A0A088A6F0 A0A2A3E370 A0A0L7R8J2 A0A158P3W4 A0A0M8ZPP8 A0A224XLJ0 A0A069DQU5 K7IYR3 C3Z3X0 A0A023F796 A0A067R164 E2BK27 A0A2J7R1W1 A0A151J8W2 R4G7S6 A0A0J7N4P1 A0A195CNY2 F4WKV5 E2AUD2 A0A2H8TIP7 A0A151WVL5 A0A195BKR7 A0A1U7Q4Z1 J9JTJ9 G3I4K4 A0A1A9YE99 A0A1A9W9Q9 A0A151JVJ1 A0A3L8DQH2 A0A154PQE2 A0A0A9WBK0 A0A026W1C0 A0A1B0AZL2 A0A1A6HW90 Q543K2 Q9WU84 A0A060YNP3 A0A0P4VXF6 A0A1U7T321 A0A0G2JSZ9 I3ME75 A0A3L7HWE1 Q9JK72 A0A0R4IHZ8 A0A0F8AKW0 F1QCS3 A0A2K8FTM4 A0A1A9VID8 A0A2K6F7Q4 D3TN65 W8C4D0 F6YJP8 L8Y595 A0A061IBV5 A0A2Y9JU61 A0A3Q2FVL3 A0A3B4DU04 A0A2K6T6L0 A0A1A7YS36 A0A1A7WRV0 A0A250YE74 B4LJV9 A0A2U9B0H9 A0A091DBB1 A0A0Q9W625 B5X3L5 A0A3Q3K2N1 A0A1A8GPI5 A0A2Y9JZY3 A0A1A8FIV1 A0A2K5RIY8 A0A1A8BXI3 A0A1A8UMW4 A0A1L8GK44 A0A1A7ZTS6 A0A1A8KT13 A0A1A8I878 A0A3G1PVI3 A0A2U3YVZ7 A0A2J8QBB0 B3MHH4 H0WLC7 A0A1A8QGN1 A0A1A8L3W1 J3KNF4 A0A1A8SF48 H2Q473 G3R2G0 A0A286ZIZ7 A0A2J8U030
A0A212ENZ9 A0A1B6G5Y1 A0A088A6F0 A0A2A3E370 A0A0L7R8J2 A0A158P3W4 A0A0M8ZPP8 A0A224XLJ0 A0A069DQU5 K7IYR3 C3Z3X0 A0A023F796 A0A067R164 E2BK27 A0A2J7R1W1 A0A151J8W2 R4G7S6 A0A0J7N4P1 A0A195CNY2 F4WKV5 E2AUD2 A0A2H8TIP7 A0A151WVL5 A0A195BKR7 A0A1U7Q4Z1 J9JTJ9 G3I4K4 A0A1A9YE99 A0A1A9W9Q9 A0A151JVJ1 A0A3L8DQH2 A0A154PQE2 A0A0A9WBK0 A0A026W1C0 A0A1B0AZL2 A0A1A6HW90 Q543K2 Q9WU84 A0A060YNP3 A0A0P4VXF6 A0A1U7T321 A0A0G2JSZ9 I3ME75 A0A3L7HWE1 Q9JK72 A0A0R4IHZ8 A0A0F8AKW0 F1QCS3 A0A2K8FTM4 A0A1A9VID8 A0A2K6F7Q4 D3TN65 W8C4D0 F6YJP8 L8Y595 A0A061IBV5 A0A2Y9JU61 A0A3Q2FVL3 A0A3B4DU04 A0A2K6T6L0 A0A1A7YS36 A0A1A7WRV0 A0A250YE74 B4LJV9 A0A2U9B0H9 A0A091DBB1 A0A0Q9W625 B5X3L5 A0A3Q3K2N1 A0A1A8GPI5 A0A2Y9JZY3 A0A1A8FIV1 A0A2K5RIY8 A0A1A8BXI3 A0A1A8UMW4 A0A1L8GK44 A0A1A7ZTS6 A0A1A8KT13 A0A1A8I878 A0A3G1PVI3 A0A2U3YVZ7 A0A2J8QBB0 B3MHH4 H0WLC7 A0A1A8QGN1 A0A1A8L3W1 J3KNF4 A0A1A8SF48 H2Q473 G3R2G0 A0A286ZIZ7 A0A2J8U030
PDB
6FP6
E-value=2.05862e-43,
Score=441
Ontologies
GO
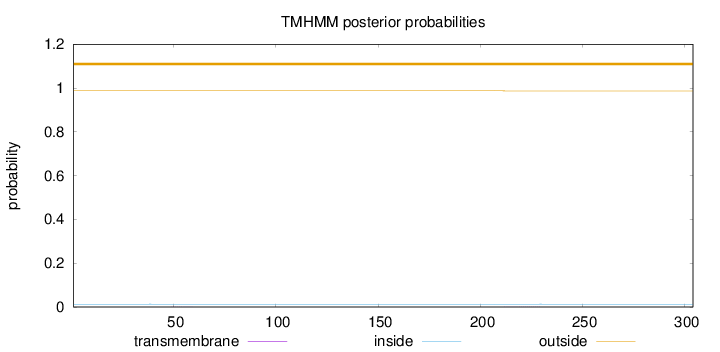
Topology
Subcellular location
Cytoplasm
Length:
304
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00539
Exp number, first 60 AAs:
0.00216
Total prob of N-in:
0.01291
outside
1 - 304
Population Genetic Test Statistics
Pi
168.045589
Theta
144.740148
Tajima's D
1.034685
CLR
0
CSRT
0.670016499175041
Interpretation
Uncertain
