Gene
KWMTBOMO06100 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001725
Annotation
PREDICTED:_cytoplasmic_aconitate_hydratase-like_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.031
Sequence
CDS
ATGGCTGGATCCACGAATCCTTACCAGAAGCTGCTGAAGACGATTGAAATTAACGGCTCTACTTACACCTACTACGATGTATCAGCCCTTGGACCTAAATACGATCGTCTCCCGTTCAGCATCAGGGTACTTCTCGAGTCATGCGTGAGGAACTGCGACAACTTCCAGGTCTTGGAGAAGGATGTCAGCAACGTCCTGGACTGGGAGTCGAAGCAGGGCAACGAGGAATCCGTGGAGATCGCCTTCAAACCTGCCAGGGTTATTTTACAGGACCTGACAGGCGTACCAGCGGTGGTAGACTTCGCGGCGATGCGTGACGCCGTGAAGGACCTGGGCGGTGACCCGGACAGGATCAACCCGATATGCCCCGCTGACCTGGTCATAGACCACTCCGTGCAGGTCGACTTCGCGAGGACACCTGACGCTCTGAACAAGAACCAGGACTTGGAGTTCGTGAGGAACAAGGAGAGGTTTCAGTTTCTGAAGTGGGGAGCGCAGGCCTTCGACAACATGCTCATAGTCCCGCCGGGCTCCGGGATCGTGCACCAAGTGAACTTGGAGTACCTGGCGCGCGTGGTGTTCACCGGCCCGCTCCTGCACCCCGACTCCGTGGTCGGCACCGACAGCCACACCACCATGATCAACGGGCTGGGAGTCGTCGGGTGGGGAGTGGGAGGCATCGAGGCGGAAGCCGTGATGCTGGGACAGGCGATCAGCATGCTGCTGCCCAAAGTCGTCGGCTACAAGCTCGTTGGAGAACTGAACCCGTTAGCCACGTCCACCGACCTGGTGCTCACGATAACGAAGCACCTGCGCTCGCTGGGCGTGGTCGGCAAGTTCGTGGAGTTCTTCGGTCCCGGGGTGGGGGCGCTGAGCATCGCCGACCGCGCCACCGTCGCCAACATGTGCCCAGAGTTCGGGGCCACACTGGCGCACTTCCCGGTCGACGCGCGCTCGCTGGAGTACCTGCACCAGACCAACCGTCCCAAAGAGAAGATCGACGTGATCGAATCGTACCTGCGCGCCACGAAGCAGTTCAGGGACTACACCAATCCCGAACAAGATCCCGTGTTCTCAGAGGTGGTGGAGCTGGACCTGGGAGACGTGGTGACGTCCGTGAGCGGGCCCAAGCGGCCCCAGGACCGCGTGTCCGTCGCCATCATGAAGAAGGACTTCCAGGACTGCTTAACCAATAAGGTCGGCTTCAAAGGCTACGGCCTGAGTCCGTCCCAGCTGTCCGCCTCCGGGGACTTCACATACAGCGACGGAAAGACTTACACCATCACCCACGGCTCTGTTATCATAGCGGCCATAACTTCGTGCACGAACACGTCCAATCCAAGCGTGATGCTCGGAGCTGGGCTCCTCGCAAAGAAAGCCGTTGAGAATGGACTATCCGTTCTTCCGTATATCAAGACATCGCTGTCTCCGGGCTCCGGAGTGGTCACGTATTATCTAAGGGAGTCTGGGGTGGTCCCGTACCTGGAGAAGCTCGGCTTCAACACGGTGGGCTACGGGTGCATGACCTGCATCGGCAACTCCGGACCCATCGACGACAACATCGCGAACACCATCGAAAAGAACGAGCTAGTCTGCTGCGGCATCCTGTCCGGGAACAGAAACTTCGAGGGCCGCATCCACCCGAACACCAGAGCCAACTACCTGGCGTCTCCGCTGCTCGTCATAGCCTACGCGCTGGCCGGGACCGTGGACGTGGACTTCGAGACCGAACCATTGGGGAAGCGCCCCGACGGCTCGGCGGTGTACCTGCGCGACATCTGGCCCACCCGCGCGGAGATACAGCGCGTCGAGAACCAGTTCGTCATCCCCGGGATGTTTAAGGAGGTGTACGCGCGGATCGAGCAGGGCTCGGCGGCGTGGCAGGCGCTGGACGTGCCGCGCGGCCGCCTGTACAGCTGGGACGCGAACTCCACCTACATCAAGAAGCCCCCATTTTTCGACGGAATGACCAAGGAGCTGCCGGCGGCGACGAGCGTGCGGGGCGCGCGCTGCCTGCTGCTGCTGGGCGACTCGGTGACCACGGATCACATCTCGCCGGCGGGATCCATCGCGCGGAACTCGCCAGCAGCGAGATACCTCGCGCAGCGCGGGCTGACAGCCAGGGAGTTCAACTCGTACGGATCTCGGCGCGGGAACGACGCGGTGATGTCCAGGGGCACGTTTGCGAACATCCGCATCGTCAACAAGATGGCGCCGGCCCCCGGACCCAAGACCACGCACCACCCCTCCGGTGACGTCATGGACATCTTCGATGCTGCCGAGAGATACGCGTCAGAGGACGTCCCCCTCATTGCCATCGTCGGGAAGGACTACGGATCCGGGTCCAGCCGGGACTGGGCGGCCAAAGGACCTTACCTCCTGGGCATAAAGGCGGTGATAGCGGAGTCGTTCGAGCGGATCCATCGCTCGAACCTGGTCGGGATGGGTCTCCTCCCGCTGCAGTTCCTGCCCGGGGAGAGCGCGGCCACGCTCGGGCTGACGGGGGCGGAGCGCTACGACATCATCCTGCCGCCCGCGCTGGCGCCCGGCCAGACCGCGACCGTACAGGTGGACAACGGCACCTCGTTCCAGGTGGTCGTACGGTTCGACACGGAAGTCGACCTGACCTACTTCAAGAACGGCGGCATCTTGAACTACATGGTGCGAAAGATGCTTTGA
Protein
MAGSTNPYQKLLKTIEINGSTYTYYDVSALGPKYDRLPFSIRVLLESCVRNCDNFQVLEKDVSNVLDWESKQGNEESVEIAFKPARVILQDLTGVPAVVDFAAMRDAVKDLGGDPDRINPICPADLVIDHSVQVDFARTPDALNKNQDLEFVRNKERFQFLKWGAQAFDNMLIVPPGSGIVHQVNLEYLARVVFTGPLLHPDSVVGTDSHTTMINGLGVVGWGVGGIEAEAVMLGQAISMLLPKVVGYKLVGELNPLATSTDLVLTITKHLRSLGVVGKFVEFFGPGVGALSIADRATVANMCPEFGATLAHFPVDARSLEYLHQTNRPKEKIDVIESYLRATKQFRDYTNPEQDPVFSEVVELDLGDVVTSVSGPKRPQDRVSVAIMKKDFQDCLTNKVGFKGYGLSPSQLSASGDFTYSDGKTYTITHGSVIIAAITSCTNTSNPSVMLGAGLLAKKAVENGLSVLPYIKTSLSPGSGVVTYYLRESGVVPYLEKLGFNTVGYGCMTCIGNSGPIDDNIANTIEKNELVCCGILSGNRNFEGRIHPNTRANYLASPLLVIAYALAGTVDVDFETEPLGKRPDGSAVYLRDIWPTRAEIQRVENQFVIPGMFKEVYARIEQGSAAWQALDVPRGRLYSWDANSTYIKKPPFFDGMTKELPAATSVRGARCLLLLGDSVTTDHISPAGSIARNSPAARYLAQRGLTAREFNSYGSRRGNDAVMSRGTFANIRIVNKMAPAPGPKTTHHPSGDVMDIFDAAERYASEDVPLIAIVGKDYGSGSSRDWAAKGPYLLGIKAVIAESFERIHRSNLVGMGLLPLQFLPGESAATLGLTGAERYDIILPPALAPGQTATVQVDNGTSFQVVVRFDTEVDLTYFKNGGILNYMVRKML
Summary
Similarity
Belongs to the aconitase/IPM isomerase family.
Uniprot
Q95UT2
A0A0N1ICI4
A0A182YMN5
A0A2M4BCZ4
A0A0T6B1Q5
A0A182KQS4
+ More
A0A182I2U1 A0A182FG79 A0A084VQU9 A0A182XLN0 A0A182P233 Q7PTD5 A0A182U6J4 A0A2M3Z2L1 A0A182UU67 A0A2M3Z2M2 A0A2M3Z2G1 A0A182G243 A0A182WJN7 A0A182J569 V5SIW4 A0A182QZY5 A0A2M3Z2H5 W5J7Y2 A0A182JTP3 A0A182MKI4 A0A336K935 A0A1Q3FHT6 B0W3V2 A0A2M4A7J8 A0A2M4A737 U5EXZ9 A0A182NKQ2 A0A182RTW6 Q16ZG5 Q6SYX7 E2I8U2 A0A224XJ57 A0A1L8E113 T1PH68 A0A1L8E0X1 B3LZR4 A0A1W4U8E0 A0A1L8E0L4 A0A171AJH3 A0A023F4H4 A0A0L0CEH4 A0A1Y1NJP8 B4K5J4 A0A1I8Q0Q1 A0A1W4V9A0 B3P1N3 B3LVY9 Q9VGZ3 B4LWE7 A0A0C9QGU2 Q9NFX2 D6WKP6 B4HIT4 C7LA95 A0A0P4VTU7 B4QV81 A0A3B0KR23 A0A0V0G5N1 A0A3L8DIG1 B4PKQ0 A0A0R3NFK1 A0A146M1J6 A0A0A9XYB8 A0A0K8SKP8 B4G695 W8C1P3 Q29AM0 B4R0Q1 Q9NFX3 Q9VCV4 A0A1A9UJQ0 B4G5W3 Q29AT7 A0A1B0G521 A0A1B0BEI3 O76934 A0A1A9XG55 A0A1A9Z9L8 B3P8K5 B4HES8 A0A1A9WY36 A0A0A1XPN2 B4NF60 B4JIK1 A0A232FEF8 E0VUE0 O76935 A0A034WDC5 A0A158NVH9 B4N9Q5 B4N8W8 A0A0K8UFW6 A0A0M4EW31 A0A0K8V3H4
A0A182I2U1 A0A182FG79 A0A084VQU9 A0A182XLN0 A0A182P233 Q7PTD5 A0A182U6J4 A0A2M3Z2L1 A0A182UU67 A0A2M3Z2M2 A0A2M3Z2G1 A0A182G243 A0A182WJN7 A0A182J569 V5SIW4 A0A182QZY5 A0A2M3Z2H5 W5J7Y2 A0A182JTP3 A0A182MKI4 A0A336K935 A0A1Q3FHT6 B0W3V2 A0A2M4A7J8 A0A2M4A737 U5EXZ9 A0A182NKQ2 A0A182RTW6 Q16ZG5 Q6SYX7 E2I8U2 A0A224XJ57 A0A1L8E113 T1PH68 A0A1L8E0X1 B3LZR4 A0A1W4U8E0 A0A1L8E0L4 A0A171AJH3 A0A023F4H4 A0A0L0CEH4 A0A1Y1NJP8 B4K5J4 A0A1I8Q0Q1 A0A1W4V9A0 B3P1N3 B3LVY9 Q9VGZ3 B4LWE7 A0A0C9QGU2 Q9NFX2 D6WKP6 B4HIT4 C7LA95 A0A0P4VTU7 B4QV81 A0A3B0KR23 A0A0V0G5N1 A0A3L8DIG1 B4PKQ0 A0A0R3NFK1 A0A146M1J6 A0A0A9XYB8 A0A0K8SKP8 B4G695 W8C1P3 Q29AM0 B4R0Q1 Q9NFX3 Q9VCV4 A0A1A9UJQ0 B4G5W3 Q29AT7 A0A1B0G521 A0A1B0BEI3 O76934 A0A1A9XG55 A0A1A9Z9L8 B3P8K5 B4HES8 A0A1A9WY36 A0A0A1XPN2 B4NF60 B4JIK1 A0A232FEF8 E0VUE0 O76935 A0A034WDC5 A0A158NVH9 B4N9Q5 B4N8W8 A0A0K8UFW6 A0A0M4EW31 A0A0K8V3H4
Pubmed
26354079
25244985
20966253
24438588
12364791
14747013
+ More
17210077 26483478 20920257 23761445 17510324 11891134 22075242 25315136 17994087 25474469 26108605 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 16679315 18362917 19820115 27129103 30249741 17550304 15632085 26823975 25401762 24495485 26109357 26109356 9660175 25830018 28648823 20566863 25348373 21347285
17210077 26483478 20920257 23761445 17510324 11891134 22075242 25315136 17994087 25474469 26108605 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 16679315 18362917 19820115 27129103 30249741 17550304 15632085 26823975 25401762 24495485 26109357 26109356 9660175 25830018 28648823 20566863 25348373 21347285
EMBL
AY032658
AAK39637.1
KQ458725
KPJ05353.1
GGFJ01001775
MBW50916.1
+ More
LJIG01016202 KRT81308.1 APCN01000180 ATLV01015326 KE525006 KFB40343.1 AAAB01008807 EAA04062.5 GGFM01002006 MBW22757.1 GGFM01002009 MBW22760.1 GGFM01001958 MBW22709.1 JXUM01039168 KQ561193 KXJ79410.1 KF647630 AHB50492.1 AXCN02000544 GGFM01001959 MBW22710.1 ADMH02002078 ETN59498.1 AXCM01004784 UFQS01000217 UFQT01000217 SSX01476.1 SSX21856.1 GFDL01007914 JAV27131.1 DS231833 EDS32249.1 GGFK01003287 MBW36608.1 GGFK01003295 MBW36616.1 GANO01000678 JAB59193.1 CH477491 EAT40025.1 AY445078 AAR15297.1 HM443949 ADN95939.1 GFTR01008253 JAW08173.1 GFDF01001899 JAV12185.1 KA647238 AFP61867.1 GFDF01001770 JAV12314.1 CH902617 EDV43058.1 GFDF01001898 JAV12186.1 GEMB01000992 JAS02153.1 GBBI01002291 JAC16421.1 JRES01000501 KNC30651.1 GEZM01001113 JAV98152.1 CH933806 EDW14031.2 CH954181 EDV49632.1 EDV41522.2 AE014297 AAF54529.1 CH940650 EDW66584.1 GBYB01013808 JAG83575.1 AY113650 AJ252017 AAM29655.1 CAB93520.1 KQ971342 EFA02996.1 CH480815 EDW42731.1 BT099645 ACV04815.1 GDKW01000650 JAI55945.1 CM000364 EDX13487.1 OUUW01000013 SPP87681.1 GECL01002806 JAP03318.1 QOIP01000007 RLU20245.1 CM000160 EDW96809.1 CM000070 KRS99928.1 GDHC01006169 JAQ12460.1 GBHO01019746 JAG23858.1 GBRD01012449 JAG53375.1 CH479179 EDW23854.1 GAMC01010961 JAB95594.1 EAL27329.3 EDX13974.1 AJ252016 CAB93519.1 AY058657 AAF56051.1 AAL13886.1 EDW23742.1 EAL27262.2 CCAG010003680 JXJN01012899 AJ223247 CAA11211.1 CH954182 EDV54100.1 EDW43239.1 GBXI01001497 JAD12795.1 CH964251 EDW82927.2 CH916369 EDV93082.1 NNAY01000342 OXU29052.1 DS235786 EEB16996.1 AJ223248 CAA11212.1 GAKP01006817 JAC52135.1 ADTU01027159 CH964232 EDW80620.2 EDW80473.2 GDHF01026765 JAI25549.1 CP012526 ALC47667.1 GDHF01018837 JAI33477.1
LJIG01016202 KRT81308.1 APCN01000180 ATLV01015326 KE525006 KFB40343.1 AAAB01008807 EAA04062.5 GGFM01002006 MBW22757.1 GGFM01002009 MBW22760.1 GGFM01001958 MBW22709.1 JXUM01039168 KQ561193 KXJ79410.1 KF647630 AHB50492.1 AXCN02000544 GGFM01001959 MBW22710.1 ADMH02002078 ETN59498.1 AXCM01004784 UFQS01000217 UFQT01000217 SSX01476.1 SSX21856.1 GFDL01007914 JAV27131.1 DS231833 EDS32249.1 GGFK01003287 MBW36608.1 GGFK01003295 MBW36616.1 GANO01000678 JAB59193.1 CH477491 EAT40025.1 AY445078 AAR15297.1 HM443949 ADN95939.1 GFTR01008253 JAW08173.1 GFDF01001899 JAV12185.1 KA647238 AFP61867.1 GFDF01001770 JAV12314.1 CH902617 EDV43058.1 GFDF01001898 JAV12186.1 GEMB01000992 JAS02153.1 GBBI01002291 JAC16421.1 JRES01000501 KNC30651.1 GEZM01001113 JAV98152.1 CH933806 EDW14031.2 CH954181 EDV49632.1 EDV41522.2 AE014297 AAF54529.1 CH940650 EDW66584.1 GBYB01013808 JAG83575.1 AY113650 AJ252017 AAM29655.1 CAB93520.1 KQ971342 EFA02996.1 CH480815 EDW42731.1 BT099645 ACV04815.1 GDKW01000650 JAI55945.1 CM000364 EDX13487.1 OUUW01000013 SPP87681.1 GECL01002806 JAP03318.1 QOIP01000007 RLU20245.1 CM000160 EDW96809.1 CM000070 KRS99928.1 GDHC01006169 JAQ12460.1 GBHO01019746 JAG23858.1 GBRD01012449 JAG53375.1 CH479179 EDW23854.1 GAMC01010961 JAB95594.1 EAL27329.3 EDX13974.1 AJ252016 CAB93519.1 AY058657 AAF56051.1 AAL13886.1 EDW23742.1 EAL27262.2 CCAG010003680 JXJN01012899 AJ223247 CAA11211.1 CH954182 EDV54100.1 EDW43239.1 GBXI01001497 JAD12795.1 CH964251 EDW82927.2 CH916369 EDV93082.1 NNAY01000342 OXU29052.1 DS235786 EEB16996.1 AJ223248 CAA11212.1 GAKP01006817 JAC52135.1 ADTU01027159 CH964232 EDW80620.2 EDW80473.2 GDHF01026765 JAI25549.1 CP012526 ALC47667.1 GDHF01018837 JAI33477.1
Proteomes
UP000053268
UP000076408
UP000075882
UP000075840
UP000069272
UP000030765
+ More
UP000076407 UP000075885 UP000007062 UP000075902 UP000075903 UP000069940 UP000249989 UP000075920 UP000075880 UP000075886 UP000000673 UP000075881 UP000075883 UP000002320 UP000075884 UP000075900 UP000008820 UP000095301 UP000007801 UP000192221 UP000037069 UP000009192 UP000095300 UP000008711 UP000000803 UP000008792 UP000007266 UP000001292 UP000000304 UP000268350 UP000279307 UP000002282 UP000001819 UP000008744 UP000078200 UP000092444 UP000092460 UP000092443 UP000092445 UP000091820 UP000007798 UP000001070 UP000215335 UP000009046 UP000005205 UP000092553
UP000076407 UP000075885 UP000007062 UP000075902 UP000075903 UP000069940 UP000249989 UP000075920 UP000075880 UP000075886 UP000000673 UP000075881 UP000075883 UP000002320 UP000075884 UP000075900 UP000008820 UP000095301 UP000007801 UP000192221 UP000037069 UP000009192 UP000095300 UP000008711 UP000000803 UP000008792 UP000007266 UP000001292 UP000000304 UP000268350 UP000279307 UP000002282 UP000001819 UP000008744 UP000078200 UP000092444 UP000092460 UP000092443 UP000092445 UP000091820 UP000007798 UP000001070 UP000215335 UP000009046 UP000005205 UP000092553
Interpro
SUPFAM
SSF53732
SSF53732
Gene 3D
ProteinModelPortal
Q95UT2
A0A0N1ICI4
A0A182YMN5
A0A2M4BCZ4
A0A0T6B1Q5
A0A182KQS4
+ More
A0A182I2U1 A0A182FG79 A0A084VQU9 A0A182XLN0 A0A182P233 Q7PTD5 A0A182U6J4 A0A2M3Z2L1 A0A182UU67 A0A2M3Z2M2 A0A2M3Z2G1 A0A182G243 A0A182WJN7 A0A182J569 V5SIW4 A0A182QZY5 A0A2M3Z2H5 W5J7Y2 A0A182JTP3 A0A182MKI4 A0A336K935 A0A1Q3FHT6 B0W3V2 A0A2M4A7J8 A0A2M4A737 U5EXZ9 A0A182NKQ2 A0A182RTW6 Q16ZG5 Q6SYX7 E2I8U2 A0A224XJ57 A0A1L8E113 T1PH68 A0A1L8E0X1 B3LZR4 A0A1W4U8E0 A0A1L8E0L4 A0A171AJH3 A0A023F4H4 A0A0L0CEH4 A0A1Y1NJP8 B4K5J4 A0A1I8Q0Q1 A0A1W4V9A0 B3P1N3 B3LVY9 Q9VGZ3 B4LWE7 A0A0C9QGU2 Q9NFX2 D6WKP6 B4HIT4 C7LA95 A0A0P4VTU7 B4QV81 A0A3B0KR23 A0A0V0G5N1 A0A3L8DIG1 B4PKQ0 A0A0R3NFK1 A0A146M1J6 A0A0A9XYB8 A0A0K8SKP8 B4G695 W8C1P3 Q29AM0 B4R0Q1 Q9NFX3 Q9VCV4 A0A1A9UJQ0 B4G5W3 Q29AT7 A0A1B0G521 A0A1B0BEI3 O76934 A0A1A9XG55 A0A1A9Z9L8 B3P8K5 B4HES8 A0A1A9WY36 A0A0A1XPN2 B4NF60 B4JIK1 A0A232FEF8 E0VUE0 O76935 A0A034WDC5 A0A158NVH9 B4N9Q5 B4N8W8 A0A0K8UFW6 A0A0M4EW31 A0A0K8V3H4
A0A182I2U1 A0A182FG79 A0A084VQU9 A0A182XLN0 A0A182P233 Q7PTD5 A0A182U6J4 A0A2M3Z2L1 A0A182UU67 A0A2M3Z2M2 A0A2M3Z2G1 A0A182G243 A0A182WJN7 A0A182J569 V5SIW4 A0A182QZY5 A0A2M3Z2H5 W5J7Y2 A0A182JTP3 A0A182MKI4 A0A336K935 A0A1Q3FHT6 B0W3V2 A0A2M4A7J8 A0A2M4A737 U5EXZ9 A0A182NKQ2 A0A182RTW6 Q16ZG5 Q6SYX7 E2I8U2 A0A224XJ57 A0A1L8E113 T1PH68 A0A1L8E0X1 B3LZR4 A0A1W4U8E0 A0A1L8E0L4 A0A171AJH3 A0A023F4H4 A0A0L0CEH4 A0A1Y1NJP8 B4K5J4 A0A1I8Q0Q1 A0A1W4V9A0 B3P1N3 B3LVY9 Q9VGZ3 B4LWE7 A0A0C9QGU2 Q9NFX2 D6WKP6 B4HIT4 C7LA95 A0A0P4VTU7 B4QV81 A0A3B0KR23 A0A0V0G5N1 A0A3L8DIG1 B4PKQ0 A0A0R3NFK1 A0A146M1J6 A0A0A9XYB8 A0A0K8SKP8 B4G695 W8C1P3 Q29AM0 B4R0Q1 Q9NFX3 Q9VCV4 A0A1A9UJQ0 B4G5W3 Q29AT7 A0A1B0G521 A0A1B0BEI3 O76934 A0A1A9XG55 A0A1A9Z9L8 B3P8K5 B4HES8 A0A1A9WY36 A0A0A1XPN2 B4NF60 B4JIK1 A0A232FEF8 E0VUE0 O76935 A0A034WDC5 A0A158NVH9 B4N9Q5 B4N8W8 A0A0K8UFW6 A0A0M4EW31 A0A0K8V3H4
PDB
2B3Y
E-value=0,
Score=3146
Ontologies
KEGG
PATHWAY
00020
Citrate cycle (TCA cycle) - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
GO
PANTHER
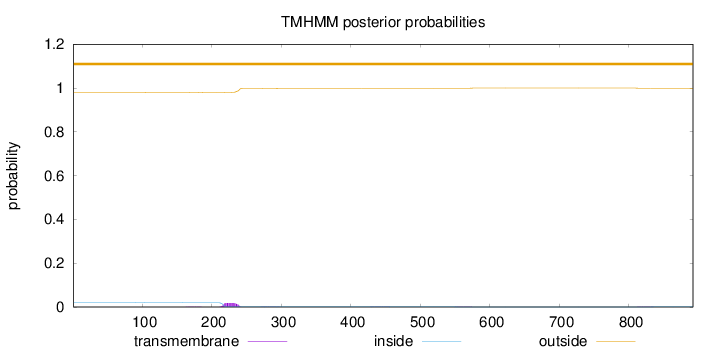
Topology
Subcellular location
Cytoplasm
Length:
892
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.47657
Exp number, first 60 AAs:
0.00114
Total prob of N-in:
0.01985
outside
1 - 892
Population Genetic Test Statistics
Pi
23.978543
Theta
162.359872
Tajima's D
1.973412
CLR
0.46464
CSRT
0.87975601219939
Interpretation
Uncertain
