Gene
KWMTBOMO06094
Pre Gene Modal
BGIBMGA001703
Annotation
PREDICTED:_lachesin-like_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.779 PlasmaMembrane Reliability : 1.234
Sequence
CDS
ATGGAGGCAGGTTGGCTGGTGGTGGTGGCGCTCTTCGTCGCTGCGAGCGCTCAGAACAATCGCAATGATGGACCACGGTTTCTGTCGAGAGGCCACTCGTTTAGGACTGTGGTTGGAGACACGCTGCTTCTTCCCTGCCAAGTCCAGAACCTTGGATCTCTGGTACTGCTGTGGAGGCGAGGCCCTGCAGTGCTCACGGCTGCGAGTCTGATGGTCACGCGAGATGAAAGGTTTCGTCTGGTCGATGGTTACAATCTTCAGATAACCGATGTCGGGCCTCAGGACGCAGGCGACTACGTTTGCCAGATATCTGATCGAGTGGCTAGGGACCAGGTGCACACTGTGGAAGTATTGGTGCCACCGAGCGTCAGAGCCTCTCCGGAGACGAGACACGTGACGGCCAGACGTGGAGGAGCTGCCGTGCTGGAGTGCCGAGCAGCTGGGAACCCTGTACCAACCGTCACGTGGCATCGATTGCTTTTCTACCAGAACGACAGCAGCTCTCGTCTCGGCGAGGGCCCGCAGCTGCAGCTCTCCCGCCTGGAGCGGCACCACGGTGGGGTCTACGCGTGCACCGTGGACAACGGCGTCGGGCCACCGGTGGTCACAGAGTTCCAGCTGCAAGTGCTGTATCCTCCAGAGATAACGGTGGAGAGATCCTGGGTTCACACTGGCGAGGGGTTCCGGGCTGAATTAGTGTGCACGGTTCTAGCCGACCCGCCCGCTGAGGTTTCTTGGTACCAGAATTCTTTTCCGCTGAGCGCTTCCGAACGCGTGACGATGTCGGCCCGAGGGAGCAACCACACTTTACTAATAGCTGATGTGCAACCCGAAGACTTCGGGAACTACAGCTGCGTGGCAGACAACAGTCTGGGTCGCGCTCGTCGTCACGTGGAGGTGTCGGGGCGGCCAGGCCCCGCACGCTTCACGTCGCCGCCGCTGGCCCGCGCGCCCCACTCCTACACGCTCAGCTTCGCCGTCGACAGCTACCCACCCCTTGATGAGTTGAGGCTACTTTACCGTCAGCTGGCCATAAACGAGTCGTTTCAACAACCAGGACGTTGGCACGACGTCGTCCTGGCGGCGCCGGCTGCCAAGGGATCATTGACGCACAGAGTCTCGCACGAGCTGCGCGGACTGCAACCCGCTGCCGTGTACGAGGCTATCGTGCAGGCGAAGAACAGATACGGGTGGAACGAGGTCAGCGACATATTCCAGTTCCACACTCTCGGCGGGCCCCACACGGCTCATGCAGACGAACTGACCCCGATGTTCTCCGCGGCGCCCGAGCCGCGACTGACCCAAAGTTTGAGTGTACTAGTCATATATACGTGTTACTGGCTGGCGTTCAGCCAATGA
Protein
MEAGWLVVVALFVAASAQNNRNDGPRFLSRGHSFRTVVGDTLLLPCQVQNLGSLVLLWRRGPAVLTAASLMVTRDERFRLVDGYNLQITDVGPQDAGDYVCQISDRVARDQVHTVEVLVPPSVRASPETRHVTARRGGAAVLECRAAGNPVPTVTWHRLLFYQNDSSSRLGEGPQLQLSRLERHHGGVYACTVDNGVGPPVVTEFQLQVLYPPEITVERSWVHTGEGFRAELVCTVLADPPAEVSWYQNSFPLSASERVTMSARGSNHTLLIADVQPEDFGNYSCVADNSLGRARRHVEVSGRPGPARFTSPPLARAPHSYTLSFAVDSYPPLDELRLLYRQLAINESFQQPGRWHDVVLAAPAAKGSLTHRVSHELRGLQPAAVYEAIVQAKNRYGWNEVSDIFQFHTLGGPHTAHADELTPMFSAAPEPRLTQSLSVLVIYTCYWLAFSQ
Summary
Uniprot
H9IWS0
A0A2H1VEA2
A0A3S2THS6
A0A212EYB7
A0A0N0PAL8
A0A0N1PJJ8
+ More
A0A2A4J2I3 A0A1S4EV00 B3LZA7 A0A1W4V8R7 B5DVY9 A0A3B0KBG7 A0A1J1HKM1 B4MB60 B4HEV3 B4R0S5 B4N9N3 Q9VCT4 A0A1B0FIK5 A0A1I8MLR2 A0A0A1X6P4 B4PMG7 B4KBD4 A0A0K8V7U5 B3P8I2 A0A1I8P5A6 W8AYC2 A0A0A9Z726 A0A0A9XXI1 A0A146LWL4 A0A1B6E7S8 Q7QBV2 P91670 Q7PSS8 A0A182Q1D4 A0A182HFW7 A0A084VGQ6 A0A336LMJ5 Q17NY0 Q7PSN2 A0A1Y9GKE3 A0A2C9GPP9 A0A1S4GEL5 A0A2C9GG51 A0A182YEC6 A0A182VFZ4 A0A182QXR3 A0A084VGS2 A0A182YEC4 A0A067RCB3 A0A182RYV8 B4MXP8 B4PDJ5 Q9VP08 A0A0J9RZL7 B3NEC1 A0A0R1E7D7 W8ASG8 A0A1J1HMF9 A0A1A9V303 A0A1B0FKK6 B3M6G5 A0A182WDE8 A0A0A1XJ55 A0A0L0BQH5 A0A3B0JZ64 A0A182U561 A0A1A9YL21 A0A1B0AR96 A0A1A9ZFZ2 A0A1I8PYR5 W5JXB6 A0A182X5I8 A0A1W4UT60 A0A182MPI2 B4LFN8 A0A0Q9WRQ9 A0A182PTK8 A0A0Q9XM42 B4KXQ7 A0A182FVU4 B4QK75 B4IAT1 A0A0M4EI05 B4J1S0 A0A0R3P897 A0A1B6KC14 A0A1J1HSR6 A0A1B6DIL5 A0A1A9WBQ4 A0A1J1J0I8 E0VBU6 A0A2C9H8B9 A0A0K8SXF0 A0A0K8SXI3 A0A0K8SX69 A0A1B6DCG8 A0A0K8SXE7 A0A0K8SYA9
A0A2A4J2I3 A0A1S4EV00 B3LZA7 A0A1W4V8R7 B5DVY9 A0A3B0KBG7 A0A1J1HKM1 B4MB60 B4HEV3 B4R0S5 B4N9N3 Q9VCT4 A0A1B0FIK5 A0A1I8MLR2 A0A0A1X6P4 B4PMG7 B4KBD4 A0A0K8V7U5 B3P8I2 A0A1I8P5A6 W8AYC2 A0A0A9Z726 A0A0A9XXI1 A0A146LWL4 A0A1B6E7S8 Q7QBV2 P91670 Q7PSS8 A0A182Q1D4 A0A182HFW7 A0A084VGQ6 A0A336LMJ5 Q17NY0 Q7PSN2 A0A1Y9GKE3 A0A2C9GPP9 A0A1S4GEL5 A0A2C9GG51 A0A182YEC6 A0A182VFZ4 A0A182QXR3 A0A084VGS2 A0A182YEC4 A0A067RCB3 A0A182RYV8 B4MXP8 B4PDJ5 Q9VP08 A0A0J9RZL7 B3NEC1 A0A0R1E7D7 W8ASG8 A0A1J1HMF9 A0A1A9V303 A0A1B0FKK6 B3M6G5 A0A182WDE8 A0A0A1XJ55 A0A0L0BQH5 A0A3B0JZ64 A0A182U561 A0A1A9YL21 A0A1B0AR96 A0A1A9ZFZ2 A0A1I8PYR5 W5JXB6 A0A182X5I8 A0A1W4UT60 A0A182MPI2 B4LFN8 A0A0Q9WRQ9 A0A182PTK8 A0A0Q9XM42 B4KXQ7 A0A182FVU4 B4QK75 B4IAT1 A0A0M4EI05 B4J1S0 A0A0R3P897 A0A1B6KC14 A0A1J1HSR6 A0A1B6DIL5 A0A1A9WBQ4 A0A1J1J0I8 E0VBU6 A0A2C9H8B9 A0A0K8SXF0 A0A0K8SXI3 A0A0K8SX69 A0A1B6DCG8 A0A0K8SXE7 A0A0K8SYA9
Pubmed
EMBL
BABH01020960
BABH01020961
ODYU01001894
SOQ38742.1
RSAL01000131
RVE46400.1
+ More
AGBW02011559 OWR46488.1 KQ458725 KPJ05378.1 KQ460140 KPJ17515.1 NWSH01003941 PCG65650.1 CH902617 EDV43034.2 CM000070 EDY67706.2 OUUW01000007 SPP83016.1 CVRI01000009 CRK88608.1 CH940656 EDW58331.2 CH480815 EDW43264.1 CM000364 EDX13998.1 CH964232 EDW80598.2 AE014297 AY060363 AAF56071.1 AAL25402.1 CCAG010005679 CCAG010005680 GBXI01007711 JAD06581.1 CM000160 EDW98072.2 CH933806 EDW16862.2 GDHF01017373 JAI34941.1 CH954182 EDV54077.1 GAMC01016705 GAMC01016704 GAMC01016703 GAMC01016702 JAB89853.1 GBHO01003340 JAG40264.1 GBHO01020031 JAG23573.1 GDHC01007070 JAQ11559.1 GEDC01003318 JAS33980.1 AAAB01008859 EAA07686.4 U78177 AAB36950.1 AAAB01008816 EAA05088.5 AXCN02001720 AXCN02001721 APCN01005430 APCN01005431 ATLV01013052 ATLV01013053 ATLV01013054 KE524837 KFB37150.1 UFQT01000020 SSX17873.1 CH477195 EAT48419.1 EAA05311.5 APCN01005433 APCN01000497 APCN01000498 APCN01000499 APCN01000500 APCN01000501 APCN01000502 AXCN02001718 AXCN02001719 ATLV01013072 ATLV01013073 KFB37166.1 KK852797 KDR16373.1 CH963876 EDW76817.2 CM000159 CH891692 EDW94994.2 KRK05177.1 AE014296 BT029930 AAF51754.3 ABM92804.1 CM002912 KMZ01059.1 CH954178 EDV52685.2 KRK05176.1 GAMC01019027 GAMC01019026 JAB87528.1 CVRI01000011 CRK89239.1 CCAG010023454 CH902618 EDV40814.1 GBXI01003372 JAD10920.1 JRES01001623 KNC21464.1 OUUW01000012 SPP87367.1 JXJN01002295 ADMH02000026 ETN68054.1 AXCM01008954 CH940647 EDW70356.2 KRF84871.1 CH933809 KRG06810.1 EDW19764.2 CM000363 EDX11412.1 CH480826 EDW44394.1 CP012525 ALC44302.1 CH916366 EDV98000.1 CH379070 KRT08479.1 GEBQ01031243 JAT08734.1 CRK89241.1 GEDC01011762 JAS25536.1 CVRI01000065 CRL05848.1 DS235042 EEB10852.1 GBRD01007922 GDHC01012121 JAG57899.1 JAQ06508.1 GBRD01007925 JAG57896.1 GBRD01007924 JAG57897.1 GEDC01013966 JAS23332.1 GBRD01007927 GDHC01018818 JAG57894.1 JAP99810.1 GBRD01007926 JAG57895.1
AGBW02011559 OWR46488.1 KQ458725 KPJ05378.1 KQ460140 KPJ17515.1 NWSH01003941 PCG65650.1 CH902617 EDV43034.2 CM000070 EDY67706.2 OUUW01000007 SPP83016.1 CVRI01000009 CRK88608.1 CH940656 EDW58331.2 CH480815 EDW43264.1 CM000364 EDX13998.1 CH964232 EDW80598.2 AE014297 AY060363 AAF56071.1 AAL25402.1 CCAG010005679 CCAG010005680 GBXI01007711 JAD06581.1 CM000160 EDW98072.2 CH933806 EDW16862.2 GDHF01017373 JAI34941.1 CH954182 EDV54077.1 GAMC01016705 GAMC01016704 GAMC01016703 GAMC01016702 JAB89853.1 GBHO01003340 JAG40264.1 GBHO01020031 JAG23573.1 GDHC01007070 JAQ11559.1 GEDC01003318 JAS33980.1 AAAB01008859 EAA07686.4 U78177 AAB36950.1 AAAB01008816 EAA05088.5 AXCN02001720 AXCN02001721 APCN01005430 APCN01005431 ATLV01013052 ATLV01013053 ATLV01013054 KE524837 KFB37150.1 UFQT01000020 SSX17873.1 CH477195 EAT48419.1 EAA05311.5 APCN01005433 APCN01000497 APCN01000498 APCN01000499 APCN01000500 APCN01000501 APCN01000502 AXCN02001718 AXCN02001719 ATLV01013072 ATLV01013073 KFB37166.1 KK852797 KDR16373.1 CH963876 EDW76817.2 CM000159 CH891692 EDW94994.2 KRK05177.1 AE014296 BT029930 AAF51754.3 ABM92804.1 CM002912 KMZ01059.1 CH954178 EDV52685.2 KRK05176.1 GAMC01019027 GAMC01019026 JAB87528.1 CVRI01000011 CRK89239.1 CCAG010023454 CH902618 EDV40814.1 GBXI01003372 JAD10920.1 JRES01001623 KNC21464.1 OUUW01000012 SPP87367.1 JXJN01002295 ADMH02000026 ETN68054.1 AXCM01008954 CH940647 EDW70356.2 KRF84871.1 CH933809 KRG06810.1 EDW19764.2 CM000363 EDX11412.1 CH480826 EDW44394.1 CP012525 ALC44302.1 CH916366 EDV98000.1 CH379070 KRT08479.1 GEBQ01031243 JAT08734.1 CRK89241.1 GEDC01011762 JAS25536.1 CVRI01000065 CRL05848.1 DS235042 EEB10852.1 GBRD01007922 GDHC01012121 JAG57899.1 JAQ06508.1 GBRD01007925 JAG57896.1 GBRD01007924 JAG57897.1 GEDC01013966 JAS23332.1 GBRD01007927 GDHC01018818 JAG57894.1 JAP99810.1 GBRD01007926 JAG57895.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000053240
UP000218220
+ More
UP000007801 UP000192221 UP000001819 UP000268350 UP000183832 UP000008792 UP000001292 UP000000304 UP000007798 UP000000803 UP000092444 UP000095301 UP000002282 UP000009192 UP000008711 UP000095300 UP000007062 UP000075886 UP000075840 UP000030765 UP000008820 UP000076408 UP000075903 UP000027135 UP000075900 UP000078200 UP000075920 UP000037069 UP000075902 UP000092443 UP000092460 UP000092445 UP000000673 UP000076407 UP000075883 UP000075885 UP000069272 UP000092553 UP000001070 UP000091820 UP000009046
UP000007801 UP000192221 UP000001819 UP000268350 UP000183832 UP000008792 UP000001292 UP000000304 UP000007798 UP000000803 UP000092444 UP000095301 UP000002282 UP000009192 UP000008711 UP000095300 UP000007062 UP000075886 UP000075840 UP000030765 UP000008820 UP000076408 UP000075903 UP000027135 UP000075900 UP000078200 UP000075920 UP000037069 UP000075902 UP000092443 UP000092460 UP000092445 UP000000673 UP000076407 UP000075883 UP000075885 UP000069272 UP000092553 UP000001070 UP000091820 UP000009046
Interpro
Gene 3D
CDD
ProteinModelPortal
H9IWS0
A0A2H1VEA2
A0A3S2THS6
A0A212EYB7
A0A0N0PAL8
A0A0N1PJJ8
+ More
A0A2A4J2I3 A0A1S4EV00 B3LZA7 A0A1W4V8R7 B5DVY9 A0A3B0KBG7 A0A1J1HKM1 B4MB60 B4HEV3 B4R0S5 B4N9N3 Q9VCT4 A0A1B0FIK5 A0A1I8MLR2 A0A0A1X6P4 B4PMG7 B4KBD4 A0A0K8V7U5 B3P8I2 A0A1I8P5A6 W8AYC2 A0A0A9Z726 A0A0A9XXI1 A0A146LWL4 A0A1B6E7S8 Q7QBV2 P91670 Q7PSS8 A0A182Q1D4 A0A182HFW7 A0A084VGQ6 A0A336LMJ5 Q17NY0 Q7PSN2 A0A1Y9GKE3 A0A2C9GPP9 A0A1S4GEL5 A0A2C9GG51 A0A182YEC6 A0A182VFZ4 A0A182QXR3 A0A084VGS2 A0A182YEC4 A0A067RCB3 A0A182RYV8 B4MXP8 B4PDJ5 Q9VP08 A0A0J9RZL7 B3NEC1 A0A0R1E7D7 W8ASG8 A0A1J1HMF9 A0A1A9V303 A0A1B0FKK6 B3M6G5 A0A182WDE8 A0A0A1XJ55 A0A0L0BQH5 A0A3B0JZ64 A0A182U561 A0A1A9YL21 A0A1B0AR96 A0A1A9ZFZ2 A0A1I8PYR5 W5JXB6 A0A182X5I8 A0A1W4UT60 A0A182MPI2 B4LFN8 A0A0Q9WRQ9 A0A182PTK8 A0A0Q9XM42 B4KXQ7 A0A182FVU4 B4QK75 B4IAT1 A0A0M4EI05 B4J1S0 A0A0R3P897 A0A1B6KC14 A0A1J1HSR6 A0A1B6DIL5 A0A1A9WBQ4 A0A1J1J0I8 E0VBU6 A0A2C9H8B9 A0A0K8SXF0 A0A0K8SXI3 A0A0K8SX69 A0A1B6DCG8 A0A0K8SXE7 A0A0K8SYA9
A0A2A4J2I3 A0A1S4EV00 B3LZA7 A0A1W4V8R7 B5DVY9 A0A3B0KBG7 A0A1J1HKM1 B4MB60 B4HEV3 B4R0S5 B4N9N3 Q9VCT4 A0A1B0FIK5 A0A1I8MLR2 A0A0A1X6P4 B4PMG7 B4KBD4 A0A0K8V7U5 B3P8I2 A0A1I8P5A6 W8AYC2 A0A0A9Z726 A0A0A9XXI1 A0A146LWL4 A0A1B6E7S8 Q7QBV2 P91670 Q7PSS8 A0A182Q1D4 A0A182HFW7 A0A084VGQ6 A0A336LMJ5 Q17NY0 Q7PSN2 A0A1Y9GKE3 A0A2C9GPP9 A0A1S4GEL5 A0A2C9GG51 A0A182YEC6 A0A182VFZ4 A0A182QXR3 A0A084VGS2 A0A182YEC4 A0A067RCB3 A0A182RYV8 B4MXP8 B4PDJ5 Q9VP08 A0A0J9RZL7 B3NEC1 A0A0R1E7D7 W8ASG8 A0A1J1HMF9 A0A1A9V303 A0A1B0FKK6 B3M6G5 A0A182WDE8 A0A0A1XJ55 A0A0L0BQH5 A0A3B0JZ64 A0A182U561 A0A1A9YL21 A0A1B0AR96 A0A1A9ZFZ2 A0A1I8PYR5 W5JXB6 A0A182X5I8 A0A1W4UT60 A0A182MPI2 B4LFN8 A0A0Q9WRQ9 A0A182PTK8 A0A0Q9XM42 B4KXQ7 A0A182FVU4 B4QK75 B4IAT1 A0A0M4EI05 B4J1S0 A0A0R3P897 A0A1B6KC14 A0A1J1HSR6 A0A1B6DIL5 A0A1A9WBQ4 A0A1J1J0I8 E0VBU6 A0A2C9H8B9 A0A0K8SXF0 A0A0K8SXI3 A0A0K8SX69 A0A1B6DCG8 A0A0K8SXE7 A0A0K8SYA9
PDB
6EG1
E-value=1.39246e-28,
Score=315
Ontologies
GO
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.976504
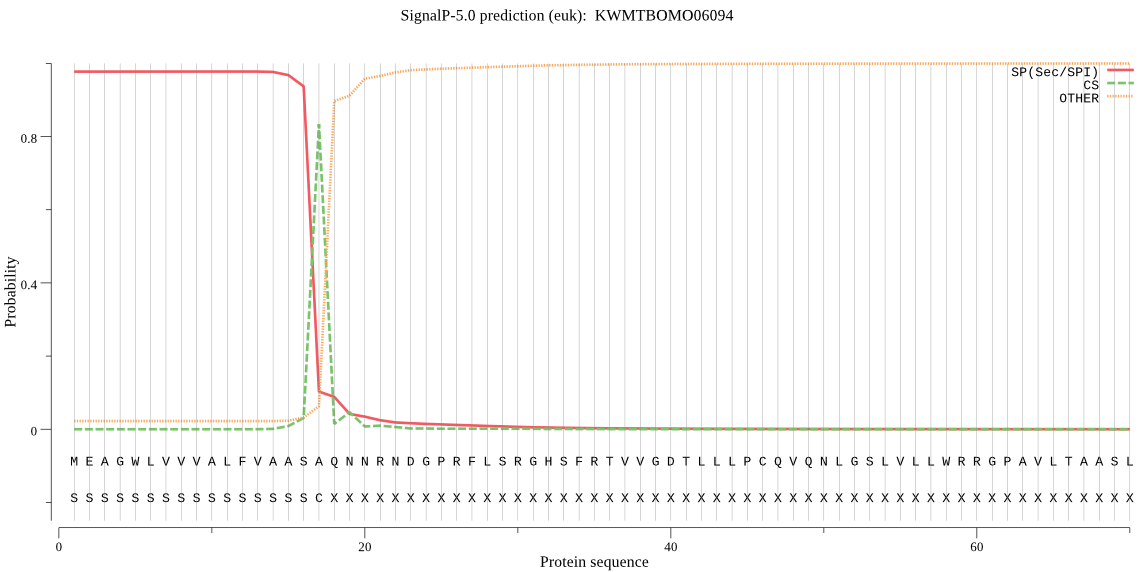
Length:
452
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.75967
Exp number, first 60 AAs:
0.25394
Total prob of N-in:
0.01581
outside
1 - 452
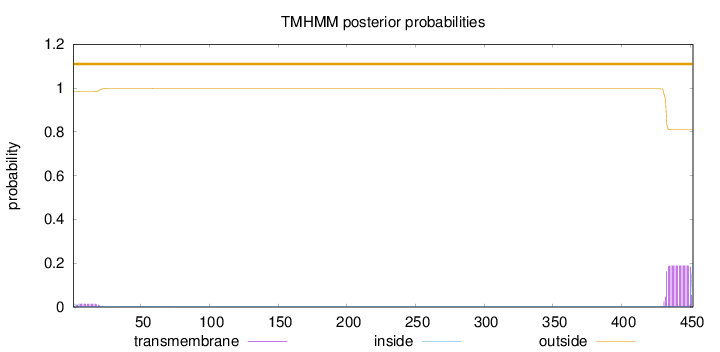
Population Genetic Test Statistics
Pi
219.895037
Theta
161.384449
Tajima's D
1.30798
CLR
0.362755
CSRT
0.744612769361532
Interpretation
Uncertain
