Gene
KWMTBOMO06088
Pre Gene Modal
BGIBMGA001721
Annotation
PREDICTED:_uncharacterized_protein_LOC101736255_isoform_X2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.959 Nuclear Reliability : 1.302
Sequence
CDS
ATGTCTCACCAGTCCACTGGCGAGATCTTCAGCAGAAATATGGAGAACCAGTCCGATAATGACGGCGTCAGACTCATTGTGCTCAACCTTCCGGAAGACGAGGGCGAGGGGCTTGGCTTCAGAGTGACGAGGACGCTGTGGGATCCGTATCCGTGGGTCCGTGAAGTGACTCCGGGGTCCAGAGCTGACGCTGCAGGCTTGAAGACCGGAGATTGTCTACTGCAAGCCGACGGAAGAGATCTCCTAGGATTACCGGTGGGACAGGTGGCCGGTCTGATCCGGGGGGACGGCGCGGGGGGCGGAGTGTCTCTCCTCGTCTGGAACTGCGGCGTCGATCCCAAGGATGATCCCGAGTTGCTGTGGAGTTGCGGCGGCGGCAGCAGGGGGGAGAGGTCACGCCGCGCGCTCGCCGGGGTGCTGAGGGCGCTCTCGTGCTGCGTCTGCGCCTCCACCGCCACCAGGGCCCTGCACTGCGCCAGATCCCATCTGTACTGCGAAAGTGTGTATCGGCAGGCTTGA
Protein
MSHQSTGEIFSRNMENQSDNDGVRLIVLNLPEDEGEGLGFRVTRTLWDPYPWVREVTPGSRADAAGLKTGDCLLQADGRDLLGLPVGQVAGLIRGDGAGGGVSLLVWNCGVDPKDDPELLWSCGGGSRGERSRRALAGVLRALSCCVCASTATRALHCARSHLYCESVYRQA
Summary
Uniprot
A0A0N1I9E2
I4DKR2
A0A0N0PAA3
A0A2H1VL52
A0A2H1VN72
A0A2W1BIU4
+ More
A0A212EWM3 A0A2A4ITJ7 D6W7S3 A0A2P8ZJY4 A0A067RKR8 E2B2V3 A0A2J7Q6D2 U4UWI5 B4GYR8 Q29AW0 Q9VCS4 E0V915 A0A1Y1MFH7 A0A182TTT9 B3LZ99 A0A0M9A9U2 W8AW39 A0A0A1XSV7 A0A034VAW7 W5JPM7 B3P8H4 B4R1C0 A0A182F6L3 A0A0K8WHR1 A0A182QA58 A0A182MSI5 A0A1W4URG2 A0A182W2P3 A0A182XVG5 A0A182NGB3 A0A1B0CRN2 A0A182XM15 A0A182R776 A0A182PUE3 Q7QBU9 A0A182HTF3 A0A182LFK8 A0A182IVD2 A0A0K8V0X9 A0A182UTH6 A0A3B0JL29 N6UDZ6 Q16YR5 B4HF95 Q16YR4 B0WCV2 A0A195B0N7 B4PN25 A0A1S4FJF9 A0A2J7Q6C8 A0A195FI34 A0A088AGE5 A0A1B6KYD5 A0A1S4FJM8 A0A1J1HQE6 A0A1B6MB84 A0A1B0B725 B4MB54 F4X757 A0A158ND94 A0A1A9XKV6 B4KBC6 A0A1B6KL69 A0A151XHI0 B4JSF0 A0A026WKE3 A0A2A3ER48 A0A310SWM4 A0A067RUH9 A0A1A9ZZR7 A0A195DJ23 A0A182VNC1 A0A0L0CLF6 A0A0J7L5X6 A0A195C2L4 E9J094 A0A154PGZ5 A0A182QCF0 B4N9F7 A0A182NGB7 A0A182U856 A0A182HTF2 A0A182XM16 A0A182T7U3 Q7PRB6 A0A182LFK7 A0A182XVG6 A0A182W2P4 A0A0L7RJQ4 A0A182LZK1 A0A336LXQ4
A0A212EWM3 A0A2A4ITJ7 D6W7S3 A0A2P8ZJY4 A0A067RKR8 E2B2V3 A0A2J7Q6D2 U4UWI5 B4GYR8 Q29AW0 Q9VCS4 E0V915 A0A1Y1MFH7 A0A182TTT9 B3LZ99 A0A0M9A9U2 W8AW39 A0A0A1XSV7 A0A034VAW7 W5JPM7 B3P8H4 B4R1C0 A0A182F6L3 A0A0K8WHR1 A0A182QA58 A0A182MSI5 A0A1W4URG2 A0A182W2P3 A0A182XVG5 A0A182NGB3 A0A1B0CRN2 A0A182XM15 A0A182R776 A0A182PUE3 Q7QBU9 A0A182HTF3 A0A182LFK8 A0A182IVD2 A0A0K8V0X9 A0A182UTH6 A0A3B0JL29 N6UDZ6 Q16YR5 B4HF95 Q16YR4 B0WCV2 A0A195B0N7 B4PN25 A0A1S4FJF9 A0A2J7Q6C8 A0A195FI34 A0A088AGE5 A0A1B6KYD5 A0A1S4FJM8 A0A1J1HQE6 A0A1B6MB84 A0A1B0B725 B4MB54 F4X757 A0A158ND94 A0A1A9XKV6 B4KBC6 A0A1B6KL69 A0A151XHI0 B4JSF0 A0A026WKE3 A0A2A3ER48 A0A310SWM4 A0A067RUH9 A0A1A9ZZR7 A0A195DJ23 A0A182VNC1 A0A0L0CLF6 A0A0J7L5X6 A0A195C2L4 E9J094 A0A154PGZ5 A0A182QCF0 B4N9F7 A0A182NGB7 A0A182U856 A0A182HTF2 A0A182XM16 A0A182T7U3 Q7PRB6 A0A182LFK7 A0A182XVG6 A0A182W2P4 A0A0L7RJQ4 A0A182LZK1 A0A336LXQ4
Pubmed
26354079
22651552
28756777
22118469
18362917
19820115
+ More
29403074 24845553 20798317 23537049 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20566863 28004739 24495485 25830018 25348373 20920257 23761445 25244985 12364791 14747013 17210077 20966253 17510324 17550304 21719571 21347285 24508170 30249741 26108605 21282665
29403074 24845553 20798317 23537049 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20566863 28004739 24495485 25830018 25348373 20920257 23761445 25244985 12364791 14747013 17210077 20966253 17510324 17550304 21719571 21347285 24508170 30249741 26108605 21282665
EMBL
KQ460140
KPJ17513.1
AK401880
BAM18502.1
KQ458725
KPJ05380.1
+ More
ODYU01003147 SOQ41531.1 ODYU01003458 SOQ42263.1 KZ150164 PZC72670.1 AGBW02011971 OWR45844.1 NWSH01007347 PCG62999.1 KQ971307 EFA11239.2 PYGN01000034 PSN56816.1 KK852411 KDR24466.1 GL445250 EFN89922.1 NEVH01017533 PNF24138.1 KB632399 ERL94630.1 CH479198 EDW28187.1 CM000070 EAL27239.2 AE014297 BT088883 AAF56081.1 ACS92855.1 DS234988 EEB09871.1 GEZM01035217 GEZM01035216 JAV83340.1 CH902617 EDV43026.1 KQ435706 KOX80157.1 GAMC01013425 GAMC01013423 JAB93130.1 GBXI01000302 JAD13990.1 GAKP01019293 JAC39659.1 ADMH02000860 ETN64835.1 CH954182 EDV54069.1 CM000364 EDX14006.1 GDHF01001687 JAI50627.1 AXCN02001622 AXCM01001961 AJWK01025003 AAAB01008859 EAA07640.4 APCN01000512 GDHF01019727 JAI32587.1 OUUW01000007 SPP83024.1 APGK01039167 APGK01039168 KB740967 ENN76882.1 CH477511 EAT39762.1 CH480815 EDW43273.1 EAT39763.1 DS231891 EDS43912.1 KQ976690 KYM78043.1 CM000160 EDW98080.1 PNF24137.1 KQ981560 KYN39927.1 GEBQ01023566 JAT16411.1 CVRI01000009 CRK88694.1 GEBQ01006843 JAT33134.1 JXJN01009349 CH940656 EDW58325.1 GL888828 EGI57669.1 ADTU01012518 CH933806 EDW16854.1 GEBQ01027765 JAT12212.1 KQ982130 KYQ59775.1 CH916373 EDV94690.1 KK107199 QOIP01000009 EZA55569.1 RLU18486.1 KZ288200 PBC33609.1 KQ759821 OAD62693.1 KDR24465.1 KQ980800 KYN12880.1 JRES01000232 KNC33130.1 LBMM01000624 KMQ97953.1 KQ978379 KYM94451.1 GL767333 EFZ13762.1 KQ434902 KZC11156.1 AXCN02001617 AXCN02001618 CH964232 EDW80590.2 EAA07526.5 KQ414581 KOC70961.1 AXCM01001380 UFQT01000265 SSX22590.1
ODYU01003147 SOQ41531.1 ODYU01003458 SOQ42263.1 KZ150164 PZC72670.1 AGBW02011971 OWR45844.1 NWSH01007347 PCG62999.1 KQ971307 EFA11239.2 PYGN01000034 PSN56816.1 KK852411 KDR24466.1 GL445250 EFN89922.1 NEVH01017533 PNF24138.1 KB632399 ERL94630.1 CH479198 EDW28187.1 CM000070 EAL27239.2 AE014297 BT088883 AAF56081.1 ACS92855.1 DS234988 EEB09871.1 GEZM01035217 GEZM01035216 JAV83340.1 CH902617 EDV43026.1 KQ435706 KOX80157.1 GAMC01013425 GAMC01013423 JAB93130.1 GBXI01000302 JAD13990.1 GAKP01019293 JAC39659.1 ADMH02000860 ETN64835.1 CH954182 EDV54069.1 CM000364 EDX14006.1 GDHF01001687 JAI50627.1 AXCN02001622 AXCM01001961 AJWK01025003 AAAB01008859 EAA07640.4 APCN01000512 GDHF01019727 JAI32587.1 OUUW01000007 SPP83024.1 APGK01039167 APGK01039168 KB740967 ENN76882.1 CH477511 EAT39762.1 CH480815 EDW43273.1 EAT39763.1 DS231891 EDS43912.1 KQ976690 KYM78043.1 CM000160 EDW98080.1 PNF24137.1 KQ981560 KYN39927.1 GEBQ01023566 JAT16411.1 CVRI01000009 CRK88694.1 GEBQ01006843 JAT33134.1 JXJN01009349 CH940656 EDW58325.1 GL888828 EGI57669.1 ADTU01012518 CH933806 EDW16854.1 GEBQ01027765 JAT12212.1 KQ982130 KYQ59775.1 CH916373 EDV94690.1 KK107199 QOIP01000009 EZA55569.1 RLU18486.1 KZ288200 PBC33609.1 KQ759821 OAD62693.1 KDR24465.1 KQ980800 KYN12880.1 JRES01000232 KNC33130.1 LBMM01000624 KMQ97953.1 KQ978379 KYM94451.1 GL767333 EFZ13762.1 KQ434902 KZC11156.1 AXCN02001617 AXCN02001618 CH964232 EDW80590.2 EAA07526.5 KQ414581 KOC70961.1 AXCM01001380 UFQT01000265 SSX22590.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000218220
UP000007266
UP000245037
+ More
UP000027135 UP000008237 UP000235965 UP000030742 UP000008744 UP000001819 UP000000803 UP000009046 UP000075902 UP000007801 UP000053105 UP000000673 UP000008711 UP000000304 UP000069272 UP000075886 UP000075883 UP000192221 UP000075920 UP000076408 UP000075884 UP000092461 UP000076407 UP000075900 UP000075885 UP000007062 UP000075840 UP000075882 UP000075880 UP000075903 UP000268350 UP000019118 UP000008820 UP000001292 UP000002320 UP000078540 UP000002282 UP000078541 UP000005203 UP000183832 UP000092460 UP000008792 UP000007755 UP000005205 UP000092443 UP000009192 UP000075809 UP000001070 UP000053097 UP000279307 UP000242457 UP000092445 UP000078492 UP000037069 UP000036403 UP000078542 UP000076502 UP000007798 UP000075901 UP000053825
UP000027135 UP000008237 UP000235965 UP000030742 UP000008744 UP000001819 UP000000803 UP000009046 UP000075902 UP000007801 UP000053105 UP000000673 UP000008711 UP000000304 UP000069272 UP000075886 UP000075883 UP000192221 UP000075920 UP000076408 UP000075884 UP000092461 UP000076407 UP000075900 UP000075885 UP000007062 UP000075840 UP000075882 UP000075880 UP000075903 UP000268350 UP000019118 UP000008820 UP000001292 UP000002320 UP000078540 UP000002282 UP000078541 UP000005203 UP000183832 UP000092460 UP000008792 UP000007755 UP000005205 UP000092443 UP000009192 UP000075809 UP000001070 UP000053097 UP000279307 UP000242457 UP000092445 UP000078492 UP000037069 UP000036403 UP000078542 UP000076502 UP000007798 UP000075901 UP000053825
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A0N1I9E2
I4DKR2
A0A0N0PAA3
A0A2H1VL52
A0A2H1VN72
A0A2W1BIU4
+ More
A0A212EWM3 A0A2A4ITJ7 D6W7S3 A0A2P8ZJY4 A0A067RKR8 E2B2V3 A0A2J7Q6D2 U4UWI5 B4GYR8 Q29AW0 Q9VCS4 E0V915 A0A1Y1MFH7 A0A182TTT9 B3LZ99 A0A0M9A9U2 W8AW39 A0A0A1XSV7 A0A034VAW7 W5JPM7 B3P8H4 B4R1C0 A0A182F6L3 A0A0K8WHR1 A0A182QA58 A0A182MSI5 A0A1W4URG2 A0A182W2P3 A0A182XVG5 A0A182NGB3 A0A1B0CRN2 A0A182XM15 A0A182R776 A0A182PUE3 Q7QBU9 A0A182HTF3 A0A182LFK8 A0A182IVD2 A0A0K8V0X9 A0A182UTH6 A0A3B0JL29 N6UDZ6 Q16YR5 B4HF95 Q16YR4 B0WCV2 A0A195B0N7 B4PN25 A0A1S4FJF9 A0A2J7Q6C8 A0A195FI34 A0A088AGE5 A0A1B6KYD5 A0A1S4FJM8 A0A1J1HQE6 A0A1B6MB84 A0A1B0B725 B4MB54 F4X757 A0A158ND94 A0A1A9XKV6 B4KBC6 A0A1B6KL69 A0A151XHI0 B4JSF0 A0A026WKE3 A0A2A3ER48 A0A310SWM4 A0A067RUH9 A0A1A9ZZR7 A0A195DJ23 A0A182VNC1 A0A0L0CLF6 A0A0J7L5X6 A0A195C2L4 E9J094 A0A154PGZ5 A0A182QCF0 B4N9F7 A0A182NGB7 A0A182U856 A0A182HTF2 A0A182XM16 A0A182T7U3 Q7PRB6 A0A182LFK7 A0A182XVG6 A0A182W2P4 A0A0L7RJQ4 A0A182LZK1 A0A336LXQ4
A0A212EWM3 A0A2A4ITJ7 D6W7S3 A0A2P8ZJY4 A0A067RKR8 E2B2V3 A0A2J7Q6D2 U4UWI5 B4GYR8 Q29AW0 Q9VCS4 E0V915 A0A1Y1MFH7 A0A182TTT9 B3LZ99 A0A0M9A9U2 W8AW39 A0A0A1XSV7 A0A034VAW7 W5JPM7 B3P8H4 B4R1C0 A0A182F6L3 A0A0K8WHR1 A0A182QA58 A0A182MSI5 A0A1W4URG2 A0A182W2P3 A0A182XVG5 A0A182NGB3 A0A1B0CRN2 A0A182XM15 A0A182R776 A0A182PUE3 Q7QBU9 A0A182HTF3 A0A182LFK8 A0A182IVD2 A0A0K8V0X9 A0A182UTH6 A0A3B0JL29 N6UDZ6 Q16YR5 B4HF95 Q16YR4 B0WCV2 A0A195B0N7 B4PN25 A0A1S4FJF9 A0A2J7Q6C8 A0A195FI34 A0A088AGE5 A0A1B6KYD5 A0A1S4FJM8 A0A1J1HQE6 A0A1B6MB84 A0A1B0B725 B4MB54 F4X757 A0A158ND94 A0A1A9XKV6 B4KBC6 A0A1B6KL69 A0A151XHI0 B4JSF0 A0A026WKE3 A0A2A3ER48 A0A310SWM4 A0A067RUH9 A0A1A9ZZR7 A0A195DJ23 A0A182VNC1 A0A0L0CLF6 A0A0J7L5X6 A0A195C2L4 E9J094 A0A154PGZ5 A0A182QCF0 B4N9F7 A0A182NGB7 A0A182U856 A0A182HTF2 A0A182XM16 A0A182T7U3 Q7PRB6 A0A182LFK7 A0A182XVG6 A0A182W2P4 A0A0L7RJQ4 A0A182LZK1 A0A336LXQ4
PDB
2LOB
E-value=0.00448807,
Score=89
Ontologies
GO
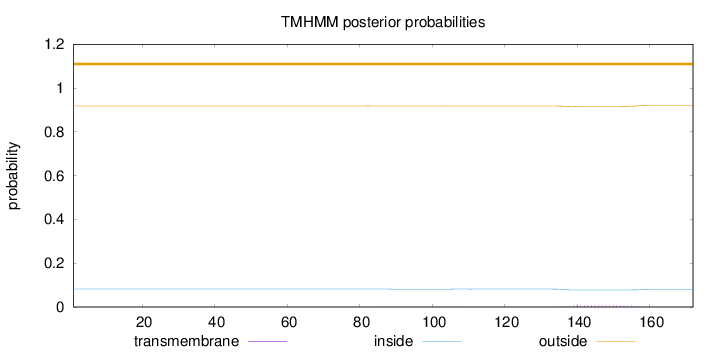
Topology
Length:
172
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13204
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08166
outside
1 - 172
Population Genetic Test Statistics
Pi
165.46896
Theta
161.929332
Tajima's D
0.758782
CLR
0.203594
CSRT
0.590770461476926
Interpretation
Uncertain
