Gene
KWMTBOMO06086 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009132
Annotation
PREDICTED:_tubulin_beta_chain-like_[Amyelois_transitella]
Full name
Tubulin beta chain
+ More
Tubulin beta-1 chain
Tubulin beta-1 chain
Alternative Name
Beta-1-tubulin
Location in the cell
Cytoplasmic Reliability : 3.502
Sequence
CDS
ATGCGTGAAATAGTTCATTTGCAGACCGGACAATGTGGCAATCAAATAGGATCGAAGTTCTGGGAAATAATCTCGGACGAGCACGGGATCGACACGACTGGACACTACCACGGAGAGAGCGACTTGCAGCTCGAGAGAATCGGTGTTTACTACAACGAAGCTGATGACGGCAAGCGCTTCGTGCCCCGCGCCGTTCTGGTGGACCTGGAGCCTGGGACCATGGACGCCATCAAGTCCTCGACGTACGGAGCGCTGTTCCGGCCGGACAACTACGTGTTTGGACAGAGCGGGGCAGGGAATAACTGGGCTAAAGGTCACTACACAGAAGGCGCTGAGCTGGTGGAATCCGTGCTGGACGTCGTCAGGAAGGAGGCCGAGGGCTGCGATTGCTTACAGGGCTTCCAGTTGACCCATTCCTTGGGAGGGGGGACGGGGTCCGGCCTTGGAACCCTCCTCCTCAGCAAACTCCGCGAGGAATACCCCGATAGGATTGTCAATACTTTCAGCGTTGTACCCTCGCCGAAGGTGTCAGATACCGTTGTGGAGCCGTACAACGCGACCCTCTCCGTGCACCAGCTGGTGGAGAACACGGACGAGACGTTCTGCATCGACAACGAGGCGCTTTACGACATCTGCTTCAGGACGTTGAGACTTGCCTCGCCCACCTACGGCGACCTCAATCACTTAGTTTCGTTGACCATGTCCGGGGTGACCACTTGTCTTCGTTTCCCGGGACAGCTGAACGCGGATCTACGGAAGCTGGCTGTTAACATGGTCCCGTTCCCGAGGCTACACTTCTTCATGCCTGGTTTCGCTCCACTGACGGCTCGTAACAGTCAAGTGTACCGCGCCCTGACCGTCCCAGAGCTGTCGCAGCAGCTGTTCAGTCCCGTGAACATGATGGCAGCCTGCGACCCTCGGCACGGGAGGTACCTCACAGTGGCCGCCATATTCCGCGGCAGGATGTCGATGAAGGAGGTTGACGAGCAGATGCTAGCCGTTCAGGACAAGAATTCTAGTTACTTCGTGGAGTGGATCCCTAACAACGTGAAGGTGGCGGTGTGCGACGTGCCGCCGCGCGGCGTCAAGATGGCCGCCACCTTCGTCGGCAACACGACCGCCATCCAAGAGATATTCAAACGGATCTCGGAGCAGTTCACGCTGATGTTCAGGAGGAAGGCGTTCCTGCACTGGTACACCGGGGAGGGGATGGACGAGATGGAGTTCACGGAGGCCGAGAGCAACATGAACGATCTCGTCTCCGAGTACCAGCAGTACGAGGAGGTTGGCGTCGACGACGGAGACTTCGAGGACCAGGAGGAAATACCAGAGGAGGACTATTTAGAAGAAGCCTAA
Protein
MREIVHLQTGQCGNQIGSKFWEIISDEHGIDTTGHYHGESDLQLERIGVYYNEADDGKRFVPRAVLVDLEPGTMDAIKSSTYGALFRPDNYVFGQSGAGNNWAKGHYTEGAELVESVLDVVRKEAEGCDCLQGFQLTHSLGGGTGSGLGTLLLSKLREEYPDRIVNTFSVVPSPKVSDTVVEPYNATLSVHQLVENTDETFCIDNEALYDICFRTLRLASPTYGDLNHLVSLTMSGVTTCLRFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTARNSQVYRALTVPELSQQLFSPVNMMAACDPRHGRYLTVAAIFRGRMSMKEVDEQMLAVQDKNSSYFVEWIPNNVKVAVCDVPPRGVKMAATFVGNTTAIQEIFKRISEQFTLMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYEEVGVDDGDFEDQEEIPEEDYLEEA
Summary
Description
Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain.
Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain (By similarity).
Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain (By similarity).
Subunit
Dimer of alpha and beta chains. A typical microtubule is a hollow water-filled tube with an outer diameter of 25 nm and an inner diameter of 15 nM. Alpha-beta heterodimers associate head-to-tail to form protofilaments running lengthwise along the microtubule wall with the beta-tubulin subunit facing the microtubule plus end conferring a structural polarity. Microtubules usually have 13 protofilaments but different protofilament numbers can be found in some organisms and specialized cells.
Dimer of alpha and beta chains. A typical microtubule is a hollow water-filled tube with an outer diameter of 25 nm and an inner diameter of 15 nM. Alpha-beta heterodimers associate head-to-tail to form protofilaments running lengthwise along the microtubule wall with the beta-tubulin subunit facing the microtubule plus end conferring a structural polarity. Microtubules usually have 13 protofilaments but different protofilament numbers can be found in some organisms and specialized cells. Interacts with mgr and Vhl.
Dimer of alpha and beta chains. A typical microtubule is a hollow water-filled tube with an outer diameter of 25 nm and an inner diameter of 15 nM. Alpha-beta heterodimers associate head-to-tail to form protofilaments running lengthwise along the microtubule wall with the beta-tubulin subunit facing the microtubule plus end conferring a structural polarity. Microtubules usually have 13 protofilaments but different protofilament numbers can be found in some organisms and specialized cells. Interacts with mgr and Vhl.
Similarity
Belongs to the tubulin family.
Keywords
Complete proteome
Cytoplasm
Cytoskeleton
GTP-binding
Microtubule
Nucleotide-binding
Phosphoprotein
Reference proteome
Feature
chain Tubulin beta-1 chain
Uniprot
A0A0N1PGL8
A0A2A4K7R7
A0A212EVB6
A0A067QYS6
A0A1V9X5G4
A0A224XCJ7
+ More
A0A0P4W3J2 A0A0V0G3G1 A0A170XZV4 A0A069DTP4 A0A226F2P2 A0A1E1XCR6 R4WCS2 A0A023GK59 G3MKF8 A0A224YT70 A0A023FK66 A0A131XPI7 A0A0K8RBZ1 A0A336MJ16 A0A1S3K224 D3PFV4 A0A0K2V4C9 A0A2J7R1W4 A0A336MPH1 A0A0V0G2I5 A0A069DZ10 R4G8S6 C1BJ29 A0A1I7YN91 K1PN21 R4G4U5 A0A3P8Z2M3 I4DMM6 A0A3S2M7U1 A0A2Z5T6R2 A0A1E1W3A9 A9YLN3 I4DIS9 A0A2M4CSY1 Q8T8B2 W5M3A6 G1KQV3 A0A1W4X306 A0A0A9Z399 A0A060X6Y9 A0A3Q3G5W8 A0A1W4VI24 A0A1L8E5H8 B4LNW8 A0A023ESD5 F6M9L9 W8BUE6 D3TR30 B4KME5 A0A1A9YJQ2 G1E6C4 A0A0G4AL61 A0A0K8V9H2 A0A1A9UYD7 T1DI49 A0A1B0ARL6 B5DUR1 A0A1Q3EUF9 B4GIN0 U5EUT8 A0A0A9Z013 A0A0A9X553 B4PB80 A0A161MFA0 A0A2J7QIE5 A0A2J7QRF5 A0A0M4EEV8 A0A1I8NLG0 B3MGF8 B4MRR0 B4HPG7 B4J625 B3NJN5 A0A0A1WMS2 A0A0P5UMT9 A0A1L8EHS1 A0A1A9ZRE2 A0A0K8TT89 A0A1A9WAB8 B4QE31 A0A0L0C7Y7 A0A0P6G935 A0A336M9C0 T1PF10 Q17GX9 Q27U48 Q24560 A0A1S3STF6 A0A2K5UIR6 A0A1Y1KNW8 A0A2R7WYX9 J3JY08 A0A131XWH3 T1DF12
A0A0P4W3J2 A0A0V0G3G1 A0A170XZV4 A0A069DTP4 A0A226F2P2 A0A1E1XCR6 R4WCS2 A0A023GK59 G3MKF8 A0A224YT70 A0A023FK66 A0A131XPI7 A0A0K8RBZ1 A0A336MJ16 A0A1S3K224 D3PFV4 A0A0K2V4C9 A0A2J7R1W4 A0A336MPH1 A0A0V0G2I5 A0A069DZ10 R4G8S6 C1BJ29 A0A1I7YN91 K1PN21 R4G4U5 A0A3P8Z2M3 I4DMM6 A0A3S2M7U1 A0A2Z5T6R2 A0A1E1W3A9 A9YLN3 I4DIS9 A0A2M4CSY1 Q8T8B2 W5M3A6 G1KQV3 A0A1W4X306 A0A0A9Z399 A0A060X6Y9 A0A3Q3G5W8 A0A1W4VI24 A0A1L8E5H8 B4LNW8 A0A023ESD5 F6M9L9 W8BUE6 D3TR30 B4KME5 A0A1A9YJQ2 G1E6C4 A0A0G4AL61 A0A0K8V9H2 A0A1A9UYD7 T1DI49 A0A1B0ARL6 B5DUR1 A0A1Q3EUF9 B4GIN0 U5EUT8 A0A0A9Z013 A0A0A9X553 B4PB80 A0A161MFA0 A0A2J7QIE5 A0A2J7QRF5 A0A0M4EEV8 A0A1I8NLG0 B3MGF8 B4MRR0 B4HPG7 B4J625 B3NJN5 A0A0A1WMS2 A0A0P5UMT9 A0A1L8EHS1 A0A1A9ZRE2 A0A0K8TT89 A0A1A9WAB8 B4QE31 A0A0L0C7Y7 A0A0P6G935 A0A336M9C0 T1PF10 Q17GX9 Q27U48 Q24560 A0A1S3STF6 A0A2K5UIR6 A0A1Y1KNW8 A0A2R7WYX9 J3JY08 A0A131XWH3 T1DF12
Pubmed
26354079
22118469
24845553
28327890
27129103
26334808
+ More
28503490 23691247 22216098 28797301 28049606 26392177 22992520 25069045 22651552 12459208 19121390 25401762 24755649 17994087 24945155 24495485 20353571 26252388 24330624 15632085 17550304 25830018 26369729 22936249 26108605 25315136 17510324 16907828 3119300 10731132 12537572 18327897 22451918 28004739 22516182
28503490 23691247 22216098 28797301 28049606 26392177 22992520 25069045 22651552 12459208 19121390 25401762 24755649 17994087 24945155 24495485 20353571 26252388 24330624 15632085 17550304 25830018 26369729 22936249 26108605 25315136 17510324 16907828 3119300 10731132 12537572 18327897 22451918 28004739 22516182
EMBL
KQ458725
KPJ05384.1
NWSH01000089
PCG79700.1
AGBW02012215
OWR45432.1
+ More
KK852818 KDR15677.1 MNPL01023288 OQR68825.1 GFTR01006309 JAW10117.1 GDKW01000582 JAI56013.1 GECL01003605 JAP02519.1 GEMB01003850 JAR99404.1 GBGD01001446 JAC87443.1 LNIX01000001 OXA63748.1 GFAC01002138 JAT97050.1 AK416954 BAN20169.1 GBBM01001101 JAC34317.1 JO842359 AEO33976.1 GFPF01006317 MAA17463.1 GBBK01002480 JAC22002.1 GEFH01000319 JAP68262.1 GADI01005146 JAA68662.1 UFQS01001300 UFQT01001300 SSX10184.1 SSX29905.1 BT120510 ADD24150.1 HACA01027819 CDW45180.1 NEVH01008202 PNF34826.1 UFQS01001981 UFQT01001981 SSX12850.1 SSX32292.1 GECL01003768 JAP02356.1 GBGD01001455 JAC87434.1 ACPB03005525 GAHY01000457 JAA77053.1 BT074608 ACO09032.1 JH817378 EKC25407.1 ACPB03021733 GAHY01000456 JAA77054.1 AK402544 BAM19166.1 RSAL01000012 RVE53452.1 LC328975 BBA78422.1 GDQN01009594 JAT81460.1 EU234504 ABY47891.1 AK401197 BAM17819.1 GGFL01004284 MBW68462.1 BABH01035358 AB072308 BAB86853.1 AHAT01021582 GBHO01005781 JAG37823.1 FR904845 CDQ72615.1 GFDF01000126 JAV13958.1 CH940648 EDW61137.1 GAPW01001430 JAC12168.1 JF767013 AEF32528.1 GAMC01001610 JAC04946.1 EZ423882 ADD20158.1 CH933808 EDW09833.1 JN029962 AEJ84083.1 KR133399 AKM70869.1 GDHF01016881 JAI35433.1 GALA01001146 JAA93706.1 JXJN01002474 CH674335 EDY71630.1 GFDL01016207 JAV18838.1 CH479183 EDW36350.1 GANO01001316 JAB58555.1 GBHO01005780 JAG37824.1 GBHO01028818 JAG14786.1 CM000158 EDW91493.1 GEMB01003849 JAR99405.1 NEVH01013629 PNF28355.1 NEVH01011896 PNF31182.1 CP012524 ALC42690.1 CH902619 EDV35701.1 CH963850 EDW74799.1 CH480816 EDW48605.1 CH916367 EDW01883.1 CH954179 EDV55213.2 GBXI01014311 JAC99980.1 GDIP01112154 JAL91560.1 GFDG01000517 JAV18282.1 GDAI01000029 JAI17574.1 CM000362 CM002911 EDX07818.1 KMY95122.1 JRES01001535 JRES01000890 KNC22195.1 KNC27524.1 GDIQ01047153 GDIQ01002074 JAN47584.1 UFQT01000630 UFQT01000929 SSX25891.1 SSX28043.1 KA647387 AFP62016.1 CH477254 EAT45945.1 DQ377071 M20419 AE013599 BT003242 AQIA01022892 GEZM01080243 JAV62278.1 KK855763 PTY23825.1 BT128131 AEE63092.1 GEFM01004769 JAP71027.1 GAMD01003291 JAA98299.1
KK852818 KDR15677.1 MNPL01023288 OQR68825.1 GFTR01006309 JAW10117.1 GDKW01000582 JAI56013.1 GECL01003605 JAP02519.1 GEMB01003850 JAR99404.1 GBGD01001446 JAC87443.1 LNIX01000001 OXA63748.1 GFAC01002138 JAT97050.1 AK416954 BAN20169.1 GBBM01001101 JAC34317.1 JO842359 AEO33976.1 GFPF01006317 MAA17463.1 GBBK01002480 JAC22002.1 GEFH01000319 JAP68262.1 GADI01005146 JAA68662.1 UFQS01001300 UFQT01001300 SSX10184.1 SSX29905.1 BT120510 ADD24150.1 HACA01027819 CDW45180.1 NEVH01008202 PNF34826.1 UFQS01001981 UFQT01001981 SSX12850.1 SSX32292.1 GECL01003768 JAP02356.1 GBGD01001455 JAC87434.1 ACPB03005525 GAHY01000457 JAA77053.1 BT074608 ACO09032.1 JH817378 EKC25407.1 ACPB03021733 GAHY01000456 JAA77054.1 AK402544 BAM19166.1 RSAL01000012 RVE53452.1 LC328975 BBA78422.1 GDQN01009594 JAT81460.1 EU234504 ABY47891.1 AK401197 BAM17819.1 GGFL01004284 MBW68462.1 BABH01035358 AB072308 BAB86853.1 AHAT01021582 GBHO01005781 JAG37823.1 FR904845 CDQ72615.1 GFDF01000126 JAV13958.1 CH940648 EDW61137.1 GAPW01001430 JAC12168.1 JF767013 AEF32528.1 GAMC01001610 JAC04946.1 EZ423882 ADD20158.1 CH933808 EDW09833.1 JN029962 AEJ84083.1 KR133399 AKM70869.1 GDHF01016881 JAI35433.1 GALA01001146 JAA93706.1 JXJN01002474 CH674335 EDY71630.1 GFDL01016207 JAV18838.1 CH479183 EDW36350.1 GANO01001316 JAB58555.1 GBHO01005780 JAG37824.1 GBHO01028818 JAG14786.1 CM000158 EDW91493.1 GEMB01003849 JAR99405.1 NEVH01013629 PNF28355.1 NEVH01011896 PNF31182.1 CP012524 ALC42690.1 CH902619 EDV35701.1 CH963850 EDW74799.1 CH480816 EDW48605.1 CH916367 EDW01883.1 CH954179 EDV55213.2 GBXI01014311 JAC99980.1 GDIP01112154 JAL91560.1 GFDG01000517 JAV18282.1 GDAI01000029 JAI17574.1 CM000362 CM002911 EDX07818.1 KMY95122.1 JRES01001535 JRES01000890 KNC22195.1 KNC27524.1 GDIQ01047153 GDIQ01002074 JAN47584.1 UFQT01000630 UFQT01000929 SSX25891.1 SSX28043.1 KA647387 AFP62016.1 CH477254 EAT45945.1 DQ377071 M20419 AE013599 BT003242 AQIA01022892 GEZM01080243 JAV62278.1 KK855763 PTY23825.1 BT128131 AEE63092.1 GEFM01004769 JAP71027.1 GAMD01003291 JAA98299.1
Proteomes
UP000053268
UP000218220
UP000007151
UP000027135
UP000192247
UP000198287
+ More
UP000085678 UP000235965 UP000015103 UP000095287 UP000005408 UP000265140 UP000283053 UP000005204 UP000018468 UP000001646 UP000192223 UP000193380 UP000261660 UP000192221 UP000008792 UP000009192 UP000092443 UP000078200 UP000092460 UP000001819 UP000008744 UP000002282 UP000092553 UP000095300 UP000007801 UP000007798 UP000001292 UP000001070 UP000008711 UP000092445 UP000091820 UP000000304 UP000037069 UP000095301 UP000008820 UP000092444 UP000000803 UP000087266 UP000233100
UP000085678 UP000235965 UP000015103 UP000095287 UP000005408 UP000265140 UP000283053 UP000005204 UP000018468 UP000001646 UP000192223 UP000193380 UP000261660 UP000192221 UP000008792 UP000009192 UP000092443 UP000078200 UP000092460 UP000001819 UP000008744 UP000002282 UP000092553 UP000095300 UP000007801 UP000007798 UP000001292 UP000001070 UP000008711 UP000092445 UP000091820 UP000000304 UP000037069 UP000095301 UP000008820 UP000092444 UP000000803 UP000087266 UP000233100
Interpro
IPR018316
Tubulin/FtsZ_2-layer-sand-dom
+ More
IPR000217 Tubulin
IPR023123 Tubulin_C
IPR036525 Tubulin/FtsZ_GTPase_sf
IPR017975 Tubulin_CS
IPR037103 Tubulin/FtsZ_C_sf
IPR002453 Beta_tubulin
IPR013838 Beta-tubulin_BS
IPR008280 Tub_FtsZ_C
IPR003008 Tubulin_FtsZ_GTPase
IPR019410 Methyltransf_16
IPR029063 SAM-dependent_MTases
IPR000217 Tubulin
IPR023123 Tubulin_C
IPR036525 Tubulin/FtsZ_GTPase_sf
IPR017975 Tubulin_CS
IPR037103 Tubulin/FtsZ_C_sf
IPR002453 Beta_tubulin
IPR013838 Beta-tubulin_BS
IPR008280 Tub_FtsZ_C
IPR003008 Tubulin_FtsZ_GTPase
IPR019410 Methyltransf_16
IPR029063 SAM-dependent_MTases
Gene 3D
ProteinModelPortal
A0A0N1PGL8
A0A2A4K7R7
A0A212EVB6
A0A067QYS6
A0A1V9X5G4
A0A224XCJ7
+ More
A0A0P4W3J2 A0A0V0G3G1 A0A170XZV4 A0A069DTP4 A0A226F2P2 A0A1E1XCR6 R4WCS2 A0A023GK59 G3MKF8 A0A224YT70 A0A023FK66 A0A131XPI7 A0A0K8RBZ1 A0A336MJ16 A0A1S3K224 D3PFV4 A0A0K2V4C9 A0A2J7R1W4 A0A336MPH1 A0A0V0G2I5 A0A069DZ10 R4G8S6 C1BJ29 A0A1I7YN91 K1PN21 R4G4U5 A0A3P8Z2M3 I4DMM6 A0A3S2M7U1 A0A2Z5T6R2 A0A1E1W3A9 A9YLN3 I4DIS9 A0A2M4CSY1 Q8T8B2 W5M3A6 G1KQV3 A0A1W4X306 A0A0A9Z399 A0A060X6Y9 A0A3Q3G5W8 A0A1W4VI24 A0A1L8E5H8 B4LNW8 A0A023ESD5 F6M9L9 W8BUE6 D3TR30 B4KME5 A0A1A9YJQ2 G1E6C4 A0A0G4AL61 A0A0K8V9H2 A0A1A9UYD7 T1DI49 A0A1B0ARL6 B5DUR1 A0A1Q3EUF9 B4GIN0 U5EUT8 A0A0A9Z013 A0A0A9X553 B4PB80 A0A161MFA0 A0A2J7QIE5 A0A2J7QRF5 A0A0M4EEV8 A0A1I8NLG0 B3MGF8 B4MRR0 B4HPG7 B4J625 B3NJN5 A0A0A1WMS2 A0A0P5UMT9 A0A1L8EHS1 A0A1A9ZRE2 A0A0K8TT89 A0A1A9WAB8 B4QE31 A0A0L0C7Y7 A0A0P6G935 A0A336M9C0 T1PF10 Q17GX9 Q27U48 Q24560 A0A1S3STF6 A0A2K5UIR6 A0A1Y1KNW8 A0A2R7WYX9 J3JY08 A0A131XWH3 T1DF12
A0A0P4W3J2 A0A0V0G3G1 A0A170XZV4 A0A069DTP4 A0A226F2P2 A0A1E1XCR6 R4WCS2 A0A023GK59 G3MKF8 A0A224YT70 A0A023FK66 A0A131XPI7 A0A0K8RBZ1 A0A336MJ16 A0A1S3K224 D3PFV4 A0A0K2V4C9 A0A2J7R1W4 A0A336MPH1 A0A0V0G2I5 A0A069DZ10 R4G8S6 C1BJ29 A0A1I7YN91 K1PN21 R4G4U5 A0A3P8Z2M3 I4DMM6 A0A3S2M7U1 A0A2Z5T6R2 A0A1E1W3A9 A9YLN3 I4DIS9 A0A2M4CSY1 Q8T8B2 W5M3A6 G1KQV3 A0A1W4X306 A0A0A9Z399 A0A060X6Y9 A0A3Q3G5W8 A0A1W4VI24 A0A1L8E5H8 B4LNW8 A0A023ESD5 F6M9L9 W8BUE6 D3TR30 B4KME5 A0A1A9YJQ2 G1E6C4 A0A0G4AL61 A0A0K8V9H2 A0A1A9UYD7 T1DI49 A0A1B0ARL6 B5DUR1 A0A1Q3EUF9 B4GIN0 U5EUT8 A0A0A9Z013 A0A0A9X553 B4PB80 A0A161MFA0 A0A2J7QIE5 A0A2J7QRF5 A0A0M4EEV8 A0A1I8NLG0 B3MGF8 B4MRR0 B4HPG7 B4J625 B3NJN5 A0A0A1WMS2 A0A0P5UMT9 A0A1L8EHS1 A0A1A9ZRE2 A0A0K8TT89 A0A1A9WAB8 B4QE31 A0A0L0C7Y7 A0A0P6G935 A0A336M9C0 T1PF10 Q17GX9 Q27U48 Q24560 A0A1S3STF6 A0A2K5UIR6 A0A1Y1KNW8 A0A2R7WYX9 J3JY08 A0A131XWH3 T1DF12
PDB
5N5N
E-value=0,
Score=1987
Ontologies
GO
PANTHER
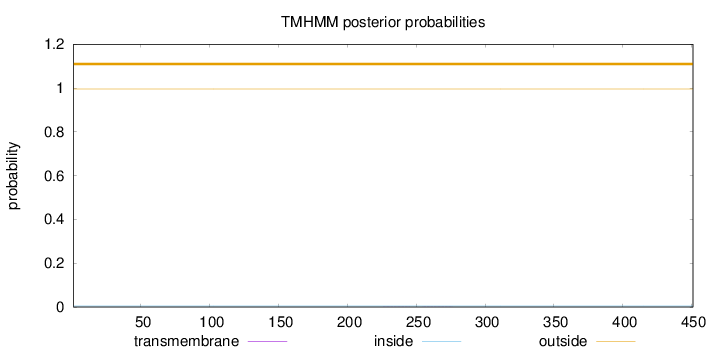
Topology
Subcellular location
Length:
451
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01126
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00449
outside
1 - 451
Population Genetic Test Statistics
Pi
179.511587
Theta
141.877449
Tajima's D
0.964132
CLR
12.658862
CSRT
0.647017649117544
Interpretation
Uncertain
