Gene
KWMTBOMO06082 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001708
Annotation
PREDICTED:_uncharacterized_protein_LOC106142738_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.653 Nuclear Reliability : 2.142
Sequence
CDS
ATGGGCAAAGATGATAAAGAAGATACAGGATTTGGATTCAAGGACAAGGCGAAGGCGGAGGACACGCTGCGGCTCCTGGAGGAGCACGACCTGAACTACAGGAGGCTGACAGTGAGAGGACTTCTCGGTAGAGCGAAGAGGGTTCTGTCAATGACGAAAGCAGAAGAGAAAATCAAAAACATCAAAGAGGCCATGGAGGTGTTCGAGAACTGGCTCGCGGACCTCGACAAGAACAAGGAGCAAAAAGAGAAGCCCGAAAAGAAAGAGAAGAAAGATGACATTGAAAAAGATGATAAAAGCGATAAGGAGTCAGATGTGGAAATGGAGGAAAAGAAAGAGAAAAAAAAGGAGCTACCCTCAGACACTGTGCCGGGCCTAGGCTTCAAGGATAAGTCTGCCGCAGAAGGGACCCTCAAAGTGTTGGACGGGAGAGACCCGGATTACCAGAGACTGGCCGTCAAGGGCCTTATAGGGAGAGCCAAGCGGGTGTTGACTTGCACCCGAGATGAGACTAAAGTATCGAACATCAAGGAGGCCATAACGGTCTTCGAGAAGTTCCTCGACGACTTCGAGAGCTTGCATCTGAGCAAAGAGAACAACCCGTACCTAAGTCTCGGCGTGGTGCGGGCCGCCGAGCAGCTGGCCGGGGAGAGCGTCACCGAGCAGCAGAAATCGTTCATAGCCGCCTACTCCTCCGTCAATGGAGAATACAAGAGGCTGAGGACCGTGGAGGAGTCGGAGGGGGGCCTCACTTGGGACATCGTTAGGAACAACGCGCTCAAACCGCTCAAAGCCACGCATGCTGAGGCGAAGCTGTTCAACGAAGAAGGCGAACCGACCCCGGAACATTTAGAGTTAATACTGTGGGCGTACTCACCGGAGGCGGCCCGCCTCAAGAAGTGCCTTCCCGAGGTGGAAACAGAAGTTAGCCGGAAGAGAAGAAGCAGCGCGCAAGAAGAGTCTCCGGCTAAAAAGAAGAAGGATTGA
Protein
MGKDDKEDTGFGFKDKAKAEDTLRLLEEHDLNYRRLTVRGLLGRAKRVLSMTKAEEKIKNIKEAMEVFENWLADLDKNKEQKEKPEKKEKKDDIEKDDKSDKESDVEMEEKKEKKKELPSDTVPGLGFKDKSAAEGTLKVLDGRDPDYQRLAVKGLIGRAKRVLTCTRDETKVSNIKEAITVFEKFLDDFESLHLSKENNPYLSLGVVRAAEQLAGESVTEQQKSFIAAYSSVNGEYKRLRTVEESEGGLTWDIVRNNALKPLKATHAEAKLFNEEGEPTPEHLELILWAYSPEAARLKKCLPEVETEVSRKRRSSAQEESPAKKKKD
Summary
Uniprot
H9IWS5
A0A3G1T1B3
A0A2W1BE38
A0A0N1IB99
A0A0N0PAA4
A0A212EVB7
+ More
A0A034WGX8 A0A182P0Q8 A0A182KAY2 A0A182HYA8 A0A182WYS6 Q7QHP0 T1PB17 A0A182VAD5 A0A182LHF9 A0A182Y6V2 A0A182NT23 A0A182TMM7 A0A0K8VVK5 A0A0A1WT71 A0A0K8TQ68 A0A0A1XEE7 A0A023EP78 A0A0L0CJ62 Q17EC1 B4PV46 A0A084VD32 B4K5J0 A0A182RL10 A0A1S4F5W8 A0A1I8QDY9 A0A182JBE3 Q29C05 B3MTV6 A0A1W4W3H9 Q7KS11 A0A182WFR2 A0A182MVD4 B4GPD8 B4HHK3 B3P6Y8 Q961C4 A0A182HBU4 B4NGP3 A0A1A9WY29 A0A3B0JFU6 A0A1B0D9B4 A0A1A9Z9K5 A0A023EN29 A0A0M3QYH0 D3TNK0 A0A1A9UJQ3 A0A1A9XG44 A0A1B0CHC2 A0A1B0BEH1 B4LWE3 B4JIK5 A0A182FLM2 A0A1Q3FLJ9 B0W2P2 A0A182Q5H7 A0A336MI20 B4QTS4
A0A034WGX8 A0A182P0Q8 A0A182KAY2 A0A182HYA8 A0A182WYS6 Q7QHP0 T1PB17 A0A182VAD5 A0A182LHF9 A0A182Y6V2 A0A182NT23 A0A182TMM7 A0A0K8VVK5 A0A0A1WT71 A0A0K8TQ68 A0A0A1XEE7 A0A023EP78 A0A0L0CJ62 Q17EC1 B4PV46 A0A084VD32 B4K5J0 A0A182RL10 A0A1S4F5W8 A0A1I8QDY9 A0A182JBE3 Q29C05 B3MTV6 A0A1W4W3H9 Q7KS11 A0A182WFR2 A0A182MVD4 B4GPD8 B4HHK3 B3P6Y8 Q961C4 A0A182HBU4 B4NGP3 A0A1A9WY29 A0A3B0JFU6 A0A1B0D9B4 A0A1A9Z9K5 A0A023EN29 A0A0M3QYH0 D3TNK0 A0A1A9UJQ3 A0A1A9XG44 A0A1B0CHC2 A0A1B0BEH1 B4LWE3 B4JIK5 A0A182FLM2 A0A1Q3FLJ9 B0W2P2 A0A182Q5H7 A0A336MI20 B4QTS4
Pubmed
EMBL
BABH01020944
MG846917
AXY94769.1
KZ150365
PZC71176.1
KQ460140
+ More
KPJ17506.1 KQ458725 KPJ05387.1 AGBW02012215 OWR45435.1 GAKP01005003 JAC53949.1 APCN01004511 AAAB01008816 EAA05227.2 KA645889 AFP60518.1 GDHF01009729 JAI42585.1 GBXI01012028 JAD02264.1 GDAI01001094 JAI16509.1 GBXI01004977 JAD09315.1 GAPW01002813 JAC10785.1 JRES01000409 KNC31519.1 CH477283 EAT44858.1 CM000160 EDW98895.1 ATLV01011135 KE524643 KFB35876.1 CH933806 EDW14027.1 CM000070 EAL26841.2 CH902623 EDV30237.1 AE014297 AAF56338.3 AGB96309.1 AHN57513.1 AXCM01002891 CH479186 EDW39021.1 CH480815 EDW43545.1 CH954182 EDV53808.1 AY051686 AAK93110.1 JXUM01032429 KQ560955 KXJ80222.1 CH964272 EDW84390.1 OUUW01000005 SPP81264.1 AJVK01004309 GAPW01002816 JAC10782.1 CP012526 ALC47663.1 CCAG010014727 EZ423002 ADD19278.1 AJWK01012206 JXJN01012899 CH940650 EDW66580.2 CH916369 EDV93086.1 GFDL01006545 JAV28500.1 DS231827 EDS29350.1 AXCN02001792 UFQT01001326 SSX30054.1 CM000364 EDX14278.1
KPJ17506.1 KQ458725 KPJ05387.1 AGBW02012215 OWR45435.1 GAKP01005003 JAC53949.1 APCN01004511 AAAB01008816 EAA05227.2 KA645889 AFP60518.1 GDHF01009729 JAI42585.1 GBXI01012028 JAD02264.1 GDAI01001094 JAI16509.1 GBXI01004977 JAD09315.1 GAPW01002813 JAC10785.1 JRES01000409 KNC31519.1 CH477283 EAT44858.1 CM000160 EDW98895.1 ATLV01011135 KE524643 KFB35876.1 CH933806 EDW14027.1 CM000070 EAL26841.2 CH902623 EDV30237.1 AE014297 AAF56338.3 AGB96309.1 AHN57513.1 AXCM01002891 CH479186 EDW39021.1 CH480815 EDW43545.1 CH954182 EDV53808.1 AY051686 AAK93110.1 JXUM01032429 KQ560955 KXJ80222.1 CH964272 EDW84390.1 OUUW01000005 SPP81264.1 AJVK01004309 GAPW01002816 JAC10782.1 CP012526 ALC47663.1 CCAG010014727 EZ423002 ADD19278.1 AJWK01012206 JXJN01012899 CH940650 EDW66580.2 CH916369 EDV93086.1 GFDL01006545 JAV28500.1 DS231827 EDS29350.1 AXCN02001792 UFQT01001326 SSX30054.1 CM000364 EDX14278.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000075885
UP000075881
+ More
UP000075840 UP000076407 UP000007062 UP000095301 UP000075903 UP000075882 UP000076408 UP000075884 UP000075902 UP000037069 UP000008820 UP000002282 UP000030765 UP000009192 UP000075900 UP000095300 UP000075880 UP000001819 UP000007801 UP000192221 UP000000803 UP000075920 UP000075883 UP000008744 UP000001292 UP000008711 UP000069940 UP000249989 UP000007798 UP000091820 UP000268350 UP000092462 UP000092445 UP000092553 UP000092444 UP000078200 UP000092443 UP000092461 UP000092460 UP000008792 UP000001070 UP000069272 UP000002320 UP000075886 UP000000304
UP000075840 UP000076407 UP000007062 UP000095301 UP000075903 UP000075882 UP000076408 UP000075884 UP000075902 UP000037069 UP000008820 UP000002282 UP000030765 UP000009192 UP000075900 UP000095300 UP000075880 UP000001819 UP000007801 UP000192221 UP000000803 UP000075920 UP000075883 UP000008744 UP000001292 UP000008711 UP000069940 UP000249989 UP000007798 UP000091820 UP000268350 UP000092462 UP000092445 UP000092553 UP000092444 UP000078200 UP000092443 UP000092461 UP000092460 UP000008792 UP000001070 UP000069272 UP000002320 UP000075886 UP000000304
Interpro
IPR026678
INO80E
ProteinModelPortal
H9IWS5
A0A3G1T1B3
A0A2W1BE38
A0A0N1IB99
A0A0N0PAA4
A0A212EVB7
+ More
A0A034WGX8 A0A182P0Q8 A0A182KAY2 A0A182HYA8 A0A182WYS6 Q7QHP0 T1PB17 A0A182VAD5 A0A182LHF9 A0A182Y6V2 A0A182NT23 A0A182TMM7 A0A0K8VVK5 A0A0A1WT71 A0A0K8TQ68 A0A0A1XEE7 A0A023EP78 A0A0L0CJ62 Q17EC1 B4PV46 A0A084VD32 B4K5J0 A0A182RL10 A0A1S4F5W8 A0A1I8QDY9 A0A182JBE3 Q29C05 B3MTV6 A0A1W4W3H9 Q7KS11 A0A182WFR2 A0A182MVD4 B4GPD8 B4HHK3 B3P6Y8 Q961C4 A0A182HBU4 B4NGP3 A0A1A9WY29 A0A3B0JFU6 A0A1B0D9B4 A0A1A9Z9K5 A0A023EN29 A0A0M3QYH0 D3TNK0 A0A1A9UJQ3 A0A1A9XG44 A0A1B0CHC2 A0A1B0BEH1 B4LWE3 B4JIK5 A0A182FLM2 A0A1Q3FLJ9 B0W2P2 A0A182Q5H7 A0A336MI20 B4QTS4
A0A034WGX8 A0A182P0Q8 A0A182KAY2 A0A182HYA8 A0A182WYS6 Q7QHP0 T1PB17 A0A182VAD5 A0A182LHF9 A0A182Y6V2 A0A182NT23 A0A182TMM7 A0A0K8VVK5 A0A0A1WT71 A0A0K8TQ68 A0A0A1XEE7 A0A023EP78 A0A0L0CJ62 Q17EC1 B4PV46 A0A084VD32 B4K5J0 A0A182RL10 A0A1S4F5W8 A0A1I8QDY9 A0A182JBE3 Q29C05 B3MTV6 A0A1W4W3H9 Q7KS11 A0A182WFR2 A0A182MVD4 B4GPD8 B4HHK3 B3P6Y8 Q961C4 A0A182HBU4 B4NGP3 A0A1A9WY29 A0A3B0JFU6 A0A1B0D9B4 A0A1A9Z9K5 A0A023EN29 A0A0M3QYH0 D3TNK0 A0A1A9UJQ3 A0A1A9XG44 A0A1B0CHC2 A0A1B0BEH1 B4LWE3 B4JIK5 A0A182FLM2 A0A1Q3FLJ9 B0W2P2 A0A182Q5H7 A0A336MI20 B4QTS4
Ontologies
PANTHER
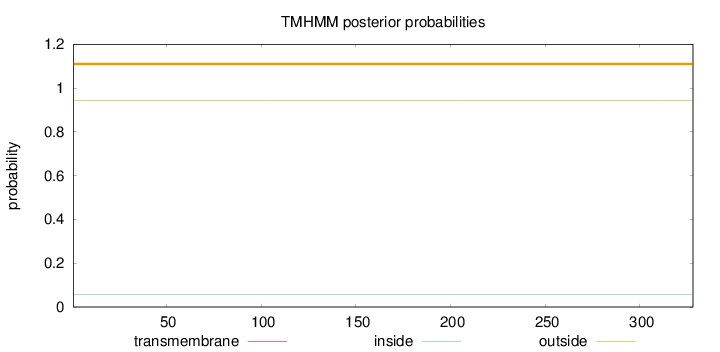
Topology
Length:
328
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05769
outside
1 - 328
Population Genetic Test Statistics
Pi
246.913527
Theta
197.37268
Tajima's D
0.805886
CLR
0.091945
CSRT
0.60161991900405
Interpretation
Uncertain
