Pre Gene Modal
BGIBMGA014518
Annotation
PREDICTED:_protein-tyrosine_sulfotransferase?_partial_[Papilio_machaon]
Full name
Protein-tyrosine sulfotransferase
Alternative Name
Transport and Golgi organization protein 13
Tyrosylprotein sulfotransferase
Tyrosylprotein sulfotransferase
Location in the cell
Mitochondrial Reliability : 1.841
Sequence
CDS
ATGTGGCGCGGCGCGCGGGCTCGGGTCGCGGCGGCGCTGGGGCTCGCCGCGCTGCTGCTGTGGGCCTGGGGCGCCGCCCCGCCACGCTCGAGCGACCGACAGGCGCCAGTCGCCGGACGCGACTTGCCGCTCATCTTCATCGGCGGCGTACCGCGCTCCGGCACCACGCTGATGCGGGCCATGCTGGACGCACATCCGGACGTGCGCTGCGGCCAGGAGACGCGCGTGGTGCCTCGCATCCTGCAGATGCGCGAGCACTGGGCCCGCTCTCAGAAAGAAAGCGTGCGTCTCGAGCAGGCCGGAGTCTCCCGCGCCGTGCTCAATAACGCCATCGCGGCCTTCTGTCTCGAGGTGATCGTGCGGCACGGCGACCCGGCACCGCGGCTCTGCAACAAAGATCCGCTCGTGCTCAAGATGGGTACGTACGTGCTGGAGCTCTTCCCGAACGCGAAGTTCCTGTTCATGGTTCGAGATGGGCGCGCCACCGTGCACTCCATCATCACAAGGAAGGTGACCATTACTGGATTCGACCTGACGAGCTACCGGCAATGTCTCACAAAGTGGAACCATGCCGTGGAACTGATGTACCAACAGTGCAAGACGATGGGATCAGACAAATGTCTTATGGTGCGCTATGAGTCGCTCGTGTTGTGGCCGCGGGAGACGATGCGACAGGTGCTTGACTTCCTGGAACTGCCCTGGGACGAGGCGGTGCTGCACCACGAGCAGTACATCAATCAGCCGAACGGAGTTGCGCTATCCAATGTGGAGCGGTCGTCGGACCAGGTGGTGCGCGCAGTGAACCGCGACGCGCTGGACAAGTGGGTGGGCGCCTTGCCGGCCGACGTGCGGGCCGACATGCCTCAGCTGGCGCCGATGCTGTCGGTGCTGGGCTACAACCCGTGGGACAATCCGCCGCCCTACCACGCGCATTCGCCGCCGCCCGCTCTCGATACCTAA
Protein
MWRGARARVAAALGLAALLLWAWGAAPPRSSDRQAPVAGRDLPLIFIGGVPRSGTTLMRAMLDAHPDVRCGQETRVVPRILQMREHWARSQKESVRLEQAGVSRAVLNNAIAAFCLEVIVRHGDPAPRLCNKDPLVLKMGTYVLELFPNAKFLFMVRDGRATVHSIITRKVTITGFDLTSYRQCLTKWNHAVELMYQQCKTMGSDKCLMVRYESLVLWPRETMRQVLDFLELPWDEAVLHHEQYINQPNGVALSNVERSSDQVVRAVNRDALDKWVGALPADVRADMPQLAPMLSVLGYNPWDNPPPYHAHSPPPALDT
Summary
Description
Catalyzes the O-sulfation of tyrosine residues within acidic motifs of polypeptides, using 3'-phosphoadenylyl sulfate (PAPS) as cosubstrate.
Catalyzes the O-sulfation of tyrosine residues within acidic motifs of polypeptides (By similarity). Has a role in protein secretion.
Catalyzes the O-sulfation of tyrosine residues within acidic motifs of polypeptides (By similarity). Has a role in protein secretion.
Catalytic Activity
3'-phosphoadenylyl sulfate + L-tyrosyl-[protein] = adenosine 3',5'-bisphosphate + H(+) + O-sulfo-L-tyrosine-[protein]
Similarity
Belongs to the protein sulfotransferase family.
Keywords
Alternative splicing
Complete proteome
Disulfide bond
Glycoprotein
Golgi apparatus
Membrane
Reference proteome
Signal-anchor
Transferase
Transmembrane
Transmembrane helix
Feature
chain Protein-tyrosine sulfotransferase
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9JY98
A0A2H1V6U6
A0A2W1BS58
A0A3S2TCT0
A0A194Q3W4
A0A2Z5UM17
+ More
A0A212FEJ6 A0A1E1WQB9 A0A0N1PFN7 A0A0L7L2Y5 T1PLM3 B4JJJ2 A0A0Q9X255 A0A1L8DW40 A0A0Q9WEA6 A0A0K8TT10 B4N2K8 S4PMJ5 Q9VYB7-2 A8JUV2 A0A1W4WE25 A0A1L8DW55 A0A0C9R5V7 A0A182Y4V7 A0A1I8MZU1 A0A182R7V3 A0A182VSP5 A0A182SXR3 A0A182M3E4 B4IG37 B4L4J9 B4M2K7 A0A182V5H3 A0A1I8NYI1 B4R4D7 M9PEH9 Q9VYB7 A0A0L0BKT2 A0A1W4W246 A0A182KK65 B3MVV9 Q7PNJ6 B4GXN1 M9PHI3 A0A084WFR8 W8BUV0 B4Q2D3 A0A182K396 A0A0M5JDU8 Q29HH7 A0A0K8WJN8 A0A182N1M5 A0A182PL19 B3NWM2 A0A182NMY5 W5JHY7 A0A2M3Z556 A0A182R032 W8B2Y3 A0A1Q3FA66 A0A182FIJ4 A0A2M4A4C1 A0A2M4BQK3 B0X4U0 A0A026WEU4 A0A232FGX3 K7IQC3 A0A182ITS5 A0A1Q3FDA8 A0A034VCM2 A0A034VE86 A0A023ESZ8 A0A0K8UPI6 A0A088AVM2 A0A195CCX8 A0A151WSP8 A0A023ER50 F4WJF5 A0A195BWD0 A0A023ER20 A0A0L7RIN8 A0A0J7L1I5 A0A023ER02 A0A2A3EK85 A0A154PE83 E2C2L6 A0A2J7R7P1 A0A2J7R7P7 A0A0N0BHR0 D6X260 A0A1Y1M665 A0A1B6J699 A0A1B6LIM7 A0A1B6LRB0 E0VHZ2 A0A067RLC6 A0A1B0Y0C2 A0A2S2NM82 A0A1B6DHI7 A0A1B6DNH9
A0A212FEJ6 A0A1E1WQB9 A0A0N1PFN7 A0A0L7L2Y5 T1PLM3 B4JJJ2 A0A0Q9X255 A0A1L8DW40 A0A0Q9WEA6 A0A0K8TT10 B4N2K8 S4PMJ5 Q9VYB7-2 A8JUV2 A0A1W4WE25 A0A1L8DW55 A0A0C9R5V7 A0A182Y4V7 A0A1I8MZU1 A0A182R7V3 A0A182VSP5 A0A182SXR3 A0A182M3E4 B4IG37 B4L4J9 B4M2K7 A0A182V5H3 A0A1I8NYI1 B4R4D7 M9PEH9 Q9VYB7 A0A0L0BKT2 A0A1W4W246 A0A182KK65 B3MVV9 Q7PNJ6 B4GXN1 M9PHI3 A0A084WFR8 W8BUV0 B4Q2D3 A0A182K396 A0A0M5JDU8 Q29HH7 A0A0K8WJN8 A0A182N1M5 A0A182PL19 B3NWM2 A0A182NMY5 W5JHY7 A0A2M3Z556 A0A182R032 W8B2Y3 A0A1Q3FA66 A0A182FIJ4 A0A2M4A4C1 A0A2M4BQK3 B0X4U0 A0A026WEU4 A0A232FGX3 K7IQC3 A0A182ITS5 A0A1Q3FDA8 A0A034VCM2 A0A034VE86 A0A023ESZ8 A0A0K8UPI6 A0A088AVM2 A0A195CCX8 A0A151WSP8 A0A023ER50 F4WJF5 A0A195BWD0 A0A023ER20 A0A0L7RIN8 A0A0J7L1I5 A0A023ER02 A0A2A3EK85 A0A154PE83 E2C2L6 A0A2J7R7P1 A0A2J7R7P7 A0A0N0BHR0 D6X260 A0A1Y1M665 A0A1B6J699 A0A1B6LIM7 A0A1B6LRB0 E0VHZ2 A0A067RLC6 A0A1B0Y0C2 A0A2S2NM82 A0A1B6DHI7 A0A1B6DNH9
EC Number
2.8.2.20
Pubmed
19121390
28756777
26354079
22118469
26227816
17994087
+ More
26369729 23622113 10731132 12537572 12537569 16452979 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 25315136 18057021 26108605 20966253 12364791 14747013 17210077 24438588 24495485 17550304 15632085 20920257 23761445 24508170 30249741 28648823 20075255 25348373 24945155 21719571 20798317 18362917 19820115 28004739 20566863 24845553
26369729 23622113 10731132 12537572 12537569 16452979 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 25315136 18057021 26108605 20966253 12364791 14747013 17210077 24438588 24495485 17550304 15632085 20920257 23761445 24508170 30249741 28648823 20075255 25348373 24945155 21719571 20798317 18362917 19820115 28004739 20566863 24845553
EMBL
BABH01042775
ODYU01000961
SOQ36506.1
KZ150039
PZC74583.1
RSAL01000349
+ More
RVE42281.1 KQ459472 KPJ00233.1 AP017499 BBB06787.1 AGBW02008947 OWR52117.1 GDQN01001876 JAT89178.1 KQ460784 KPJ12205.1 JTDY01003272 KOB69832.1 KA648838 AFP63467.1 CH916370 EDV99744.1 CH963925 KRF98914.1 GFDF01003438 JAV10646.1 CH940651 KRF82470.1 GDAI01000332 JAI17271.1 EDW78597.2 GAIX01003520 JAA89040.1 AY124548 AE014298 AY119073 BT021950 ABW09407.1 GFDF01003407 JAV10677.1 GBYB01003335 JAG73102.1 AXCM01011898 CH480835 EDW48771.1 CH933810 EDW07477.2 EDW65911.1 KRF82471.1 CM000366 EDX17839.1 AGB95362.1 JRES01001705 KNC20700.1 CH902625 EDV35104.2 AAAB01008963 EAA12079.6 CH479196 EDW27508.1 AGB95360.1 ATLV01023392 KE525342 KFB49062.1 GAMC01013646 JAB92909.1 CM000162 EDX01594.2 CP012528 ALC49544.1 CH379064 EAL31781.3 GDHF01000941 JAI51373.1 CH954180 EDV47184.2 ADMH02001203 ETN63741.1 GGFM01002895 MBW23646.1 AXCN02001440 GAMC01013644 GAMC01013643 JAB92912.1 GFDL01010647 JAV24398.1 GGFK01002322 MBW35643.1 GGFJ01006112 MBW55253.1 DS232354 EDS40555.1 KK107260 QOIP01000001 EZA54181.1 RLU27219.1 NNAY01000253 OXU29689.1 GFDL01009563 JAV25482.1 GAKP01019085 GAKP01019083 JAC39867.1 GAKP01019086 GAKP01019084 JAC39868.1 GAPW01002134 JAC11464.1 GDHF01023717 JAI28597.1 KQ978023 KYM97938.1 KQ982769 KYQ50874.1 GAPW01001968 JAC11630.1 GL888182 EGI65733.1 KQ976401 KYM92590.1 GAPW01002003 JAC11595.1 KQ414582 KOC70832.1 LBMM01001421 KMQ96354.1 GAPW01002028 JAC11570.1 KZ288220 PBC32185.1 KQ434874 KZC09734.1 GL452167 EFN77815.1 NEVH01006727 PNF36855.1 PNF36853.1 KQ435745 KOX76592.1 KQ971371 EFA09909.1 GEZM01039769 JAV81214.1 GECU01012999 JAS94707.1 GEBQ01016435 JAT23542.1 GEBQ01013800 JAT26177.1 DS235172 EEB12998.1 KK852556 KDR21420.1 KT000409 ANI86007.1 GGMR01005671 MBY18290.1 GEDC01029721 GEDC01012218 GEDC01011482 GEDC01003081 JAS07577.1 JAS25080.1 JAS25816.1 JAS34217.1 GEDC01014830 GEDC01010076 JAS22468.1 JAS27222.1
RVE42281.1 KQ459472 KPJ00233.1 AP017499 BBB06787.1 AGBW02008947 OWR52117.1 GDQN01001876 JAT89178.1 KQ460784 KPJ12205.1 JTDY01003272 KOB69832.1 KA648838 AFP63467.1 CH916370 EDV99744.1 CH963925 KRF98914.1 GFDF01003438 JAV10646.1 CH940651 KRF82470.1 GDAI01000332 JAI17271.1 EDW78597.2 GAIX01003520 JAA89040.1 AY124548 AE014298 AY119073 BT021950 ABW09407.1 GFDF01003407 JAV10677.1 GBYB01003335 JAG73102.1 AXCM01011898 CH480835 EDW48771.1 CH933810 EDW07477.2 EDW65911.1 KRF82471.1 CM000366 EDX17839.1 AGB95362.1 JRES01001705 KNC20700.1 CH902625 EDV35104.2 AAAB01008963 EAA12079.6 CH479196 EDW27508.1 AGB95360.1 ATLV01023392 KE525342 KFB49062.1 GAMC01013646 JAB92909.1 CM000162 EDX01594.2 CP012528 ALC49544.1 CH379064 EAL31781.3 GDHF01000941 JAI51373.1 CH954180 EDV47184.2 ADMH02001203 ETN63741.1 GGFM01002895 MBW23646.1 AXCN02001440 GAMC01013644 GAMC01013643 JAB92912.1 GFDL01010647 JAV24398.1 GGFK01002322 MBW35643.1 GGFJ01006112 MBW55253.1 DS232354 EDS40555.1 KK107260 QOIP01000001 EZA54181.1 RLU27219.1 NNAY01000253 OXU29689.1 GFDL01009563 JAV25482.1 GAKP01019085 GAKP01019083 JAC39867.1 GAKP01019086 GAKP01019084 JAC39868.1 GAPW01002134 JAC11464.1 GDHF01023717 JAI28597.1 KQ978023 KYM97938.1 KQ982769 KYQ50874.1 GAPW01001968 JAC11630.1 GL888182 EGI65733.1 KQ976401 KYM92590.1 GAPW01002003 JAC11595.1 KQ414582 KOC70832.1 LBMM01001421 KMQ96354.1 GAPW01002028 JAC11570.1 KZ288220 PBC32185.1 KQ434874 KZC09734.1 GL452167 EFN77815.1 NEVH01006727 PNF36855.1 PNF36853.1 KQ435745 KOX76592.1 KQ971371 EFA09909.1 GEZM01039769 JAV81214.1 GECU01012999 JAS94707.1 GEBQ01016435 JAT23542.1 GEBQ01013800 JAT26177.1 DS235172 EEB12998.1 KK852556 KDR21420.1 KT000409 ANI86007.1 GGMR01005671 MBY18290.1 GEDC01029721 GEDC01012218 GEDC01011482 GEDC01003081 JAS07577.1 JAS25080.1 JAS25816.1 JAS34217.1 GEDC01014830 GEDC01010076 JAS22468.1 JAS27222.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000007151
UP000053240
UP000037510
+ More
UP000001070 UP000007798 UP000008792 UP000000803 UP000192221 UP000076408 UP000095301 UP000075900 UP000075920 UP000075901 UP000075883 UP000001292 UP000009192 UP000075903 UP000095300 UP000000304 UP000037069 UP000075882 UP000007801 UP000007062 UP000008744 UP000030765 UP000002282 UP000075881 UP000092553 UP000001819 UP000075884 UP000075885 UP000008711 UP000000673 UP000075886 UP000069272 UP000002320 UP000053097 UP000279307 UP000215335 UP000002358 UP000075880 UP000005203 UP000078542 UP000075809 UP000007755 UP000078540 UP000053825 UP000036403 UP000242457 UP000076502 UP000008237 UP000235965 UP000053105 UP000007266 UP000009046 UP000027135
UP000001070 UP000007798 UP000008792 UP000000803 UP000192221 UP000076408 UP000095301 UP000075900 UP000075920 UP000075901 UP000075883 UP000001292 UP000009192 UP000075903 UP000095300 UP000000304 UP000037069 UP000075882 UP000007801 UP000007062 UP000008744 UP000030765 UP000002282 UP000075881 UP000092553 UP000001819 UP000075884 UP000075885 UP000008711 UP000000673 UP000075886 UP000069272 UP000002320 UP000053097 UP000279307 UP000215335 UP000002358 UP000075880 UP000005203 UP000078542 UP000075809 UP000007755 UP000078540 UP000053825 UP000036403 UP000242457 UP000076502 UP000008237 UP000235965 UP000053105 UP000007266 UP000009046 UP000027135
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9JY98
A0A2H1V6U6
A0A2W1BS58
A0A3S2TCT0
A0A194Q3W4
A0A2Z5UM17
+ More
A0A212FEJ6 A0A1E1WQB9 A0A0N1PFN7 A0A0L7L2Y5 T1PLM3 B4JJJ2 A0A0Q9X255 A0A1L8DW40 A0A0Q9WEA6 A0A0K8TT10 B4N2K8 S4PMJ5 Q9VYB7-2 A8JUV2 A0A1W4WE25 A0A1L8DW55 A0A0C9R5V7 A0A182Y4V7 A0A1I8MZU1 A0A182R7V3 A0A182VSP5 A0A182SXR3 A0A182M3E4 B4IG37 B4L4J9 B4M2K7 A0A182V5H3 A0A1I8NYI1 B4R4D7 M9PEH9 Q9VYB7 A0A0L0BKT2 A0A1W4W246 A0A182KK65 B3MVV9 Q7PNJ6 B4GXN1 M9PHI3 A0A084WFR8 W8BUV0 B4Q2D3 A0A182K396 A0A0M5JDU8 Q29HH7 A0A0K8WJN8 A0A182N1M5 A0A182PL19 B3NWM2 A0A182NMY5 W5JHY7 A0A2M3Z556 A0A182R032 W8B2Y3 A0A1Q3FA66 A0A182FIJ4 A0A2M4A4C1 A0A2M4BQK3 B0X4U0 A0A026WEU4 A0A232FGX3 K7IQC3 A0A182ITS5 A0A1Q3FDA8 A0A034VCM2 A0A034VE86 A0A023ESZ8 A0A0K8UPI6 A0A088AVM2 A0A195CCX8 A0A151WSP8 A0A023ER50 F4WJF5 A0A195BWD0 A0A023ER20 A0A0L7RIN8 A0A0J7L1I5 A0A023ER02 A0A2A3EK85 A0A154PE83 E2C2L6 A0A2J7R7P1 A0A2J7R7P7 A0A0N0BHR0 D6X260 A0A1Y1M665 A0A1B6J699 A0A1B6LIM7 A0A1B6LRB0 E0VHZ2 A0A067RLC6 A0A1B0Y0C2 A0A2S2NM82 A0A1B6DHI7 A0A1B6DNH9
A0A212FEJ6 A0A1E1WQB9 A0A0N1PFN7 A0A0L7L2Y5 T1PLM3 B4JJJ2 A0A0Q9X255 A0A1L8DW40 A0A0Q9WEA6 A0A0K8TT10 B4N2K8 S4PMJ5 Q9VYB7-2 A8JUV2 A0A1W4WE25 A0A1L8DW55 A0A0C9R5V7 A0A182Y4V7 A0A1I8MZU1 A0A182R7V3 A0A182VSP5 A0A182SXR3 A0A182M3E4 B4IG37 B4L4J9 B4M2K7 A0A182V5H3 A0A1I8NYI1 B4R4D7 M9PEH9 Q9VYB7 A0A0L0BKT2 A0A1W4W246 A0A182KK65 B3MVV9 Q7PNJ6 B4GXN1 M9PHI3 A0A084WFR8 W8BUV0 B4Q2D3 A0A182K396 A0A0M5JDU8 Q29HH7 A0A0K8WJN8 A0A182N1M5 A0A182PL19 B3NWM2 A0A182NMY5 W5JHY7 A0A2M3Z556 A0A182R032 W8B2Y3 A0A1Q3FA66 A0A182FIJ4 A0A2M4A4C1 A0A2M4BQK3 B0X4U0 A0A026WEU4 A0A232FGX3 K7IQC3 A0A182ITS5 A0A1Q3FDA8 A0A034VCM2 A0A034VE86 A0A023ESZ8 A0A0K8UPI6 A0A088AVM2 A0A195CCX8 A0A151WSP8 A0A023ER50 F4WJF5 A0A195BWD0 A0A023ER20 A0A0L7RIN8 A0A0J7L1I5 A0A023ER02 A0A2A3EK85 A0A154PE83 E2C2L6 A0A2J7R7P1 A0A2J7R7P7 A0A0N0BHR0 D6X260 A0A1Y1M665 A0A1B6J699 A0A1B6LIM7 A0A1B6LRB0 E0VHZ2 A0A067RLC6 A0A1B0Y0C2 A0A2S2NM82 A0A1B6DHI7 A0A1B6DNH9
PDB
5WRJ
E-value=3.93733e-104,
Score=965
Ontologies
PANTHER
Topology
Subcellular location
Golgi apparatus membrane
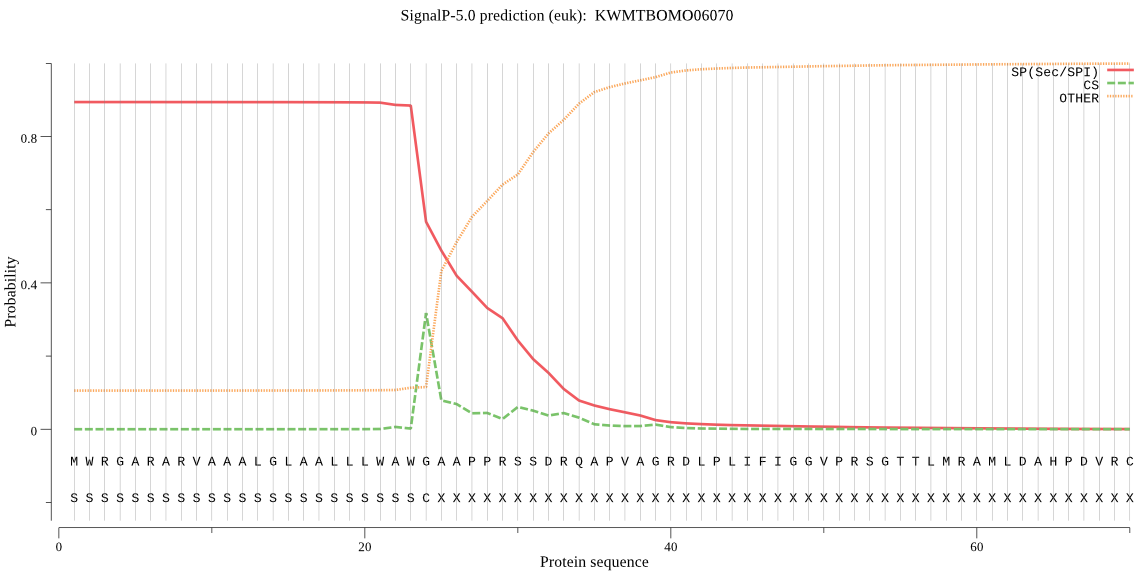
SignalP
Position: 1 - 24,
Likelihood: 0.894201
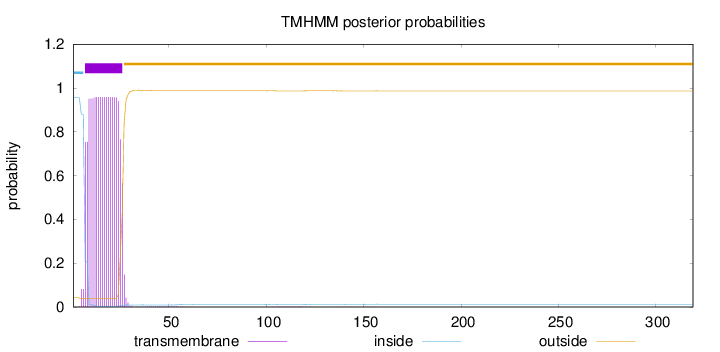
Length:
319
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.63141
Exp number, first 60 AAs:
18.58278
Total prob of N-in:
0.95659
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 26
outside
27 - 319
Population Genetic Test Statistics
Pi
2.263482
Theta
5.984251
Tajima's D
-1.799661
CLR
0.072022
CSRT
0.0254987250637468
Interpretation
Uncertain
