Gene
KWMTBOMO06069
Pre Gene Modal
BGIBMGA002851
Annotation
PREDICTED:_probable_G-protein_coupled_receptor_Mth-like_1_[Amyelois_transitella]
Full name
Probable G-protein coupled receptor Mth-like 1
Alternative Name
Protein methuselah-like 1
Location in the cell
PlasmaMembrane Reliability : 3.907
Sequence
CDS
ATGCTGGCGACTCTCGGCGTCGTGGTCGCGTGGACAGTGTGCTGGGTGTCAGCGGGCGGCGCGCACTCCCAACCTCTCTCTATTCCGCGCTGCTGTCCCGCCGGCCTCAGCCTCGATCTTGCGGCGCTCAGTGCGCTCGTGATCACCCCCGCCGCTGTGCCCTCGGCCTGTGTGCCCCAGACTGATCTTCAGCAAAAATGGGTACCGCTCGTGTACGGGCCGGTCGTGGGACGCTTCTTGGAGCGTGGTCGTTTGCCCGCACAGTGGAAGTTTCGCGACGCGACCGTCCCGAATTGCAAGGAGCTCCAGATCATTCCAGAACACGCGGCTGCTTACGCGCTTCTCAGAAATGACGGCTCTCTAATGATGATCCGGAGTAGAGGACGAGCGCTGGCCTTGTCGCGATACTGCACGGATGCCGCTGCAGCCCTCGTGTGCGCCGAGGATGTCACGCACACGTTATCCAAGTGTTGTGGCGAGGGACGAGTGTTTGACGGTGGCCATTGTATCTTGAACGAGTCCAAGTCTGTTGAGGTTCTTGCGGAACTGCGGAAATTAGCTAACGGCACCGTCGTCGGAGGCGGGTGGCCCACATGCGTCGCCGGCTTGAGCTACGGGGTGACGGGCGCCCTGAGCGGAGCCGTGCTGATGCCGGACGGCGCGCTAGAACTTAGTGGCACCAAGCTAGAGGCGGGTGCGTGGTGTGCAGAAGCAGTGGTGGGTGAGGAGGGCACGCGCATTTTGACTTGCGAAGCGGTGGCGAGCGCGGTGGTCCCGCGGCGGACAGCTCGGCACGCGCTCTACGGTGCAGGGCTGGCTGTGGGCGCGGCGTTCCTGGCCGCCACGCTGGCGGCTGGATTCGCGCTGCCGGCGGCTCACCACGCGCTGCACTGGCGGTGCCAGACGCACTATGTGGCGGCGCTGATGCTGGGCGACGTGCTGCTGGCGGCGACGCAGCTGGCCGACAACTCCGTGCCGCCGACATTGTGTCGCGCGCTAGCGGTCTGCATGCACTTCTTGTTCCTGTCCGCGTTCTTCTGGCTCAACACGATGTGTTTCAACATTTGGTGGACGTTCCGGGACTTCCGCCCGACCTCGCTGGAACGCGGCCAGGAGGCGTGGCGCCTCCGCGTGTACATGGTGTACGCGTGGGGCGGGCCGCTGCTGGTGGCCGGCACCGGCGCCCTGCTCGACCACCTGCCGGCCGGCGCCGCGCCTCTGCTGCGGCCGCGCTTCGCCGTGCAGCGTTGCTGGTTTTATGGTGACATGGAGATTCTCGTCTACTTCTTCGGTCCTGTGGGCGTCTTGCTTCTGGTCAATCTTGCACTCTTCATTTCTACCACGCGCCAGCTGACCTGCGGCCTCTGGCGGCGCGATGAGGTCAAGTCTACATCCGAGAGGGCAGCTTTGGGACGCGTGTGTGCCAAGCTGGTAGTAGTGATGGGCGTGACGTGGGGTGCGGATGTGGTGTCGTGGGCGGCGGGCGGGCCGGAGTACGTGTGGTACGCGACGGATCTGCTCAATGCTCTCCAGGGCGTGCTCATCTTCCTGGTGGTGGGGTGTCAACCGCACGCCTGGGCAGCGCTGAAGCGGTGTGCTGCGGCGTGCTGCGCGAAGGGCGGCAGCGGAGGCGGAGAGCGGGGCGCGGGCCGCCACTCCTCGACGCACCTGCCGTCCTGTGGGGAATCTCTGACGCACACCACGCAGGCCGCCGCCCCGGCCCCTGCGCCAGTACCTACACCCCGCGTCCCCATGGAGACGGTCTGCTGA
Protein
MLATLGVVVAWTVCWVSAGGAHSQPLSIPRCCPAGLSLDLAALSALVITPAAVPSACVPQTDLQQKWVPLVYGPVVGRFLERGRLPAQWKFRDATVPNCKELQIIPEHAAAYALLRNDGSLMMIRSRGRALALSRYCTDAAAALVCAEDVTHTLSKCCGEGRVFDGGHCILNESKSVEVLAELRKLANGTVVGGGWPTCVAGLSYGVTGALSGAVLMPDGALELSGTKLEAGAWCAEAVVGEEGTRILTCEAVASAVVPRRTARHALYGAGLAVGAAFLAATLAAGFALPAAHHALHWRCQTHYVAALMLGDVLLAATQLADNSVPPTLCRALAVCMHFLFLSAFFWLNTMCFNIWWTFRDFRPTSLERGQEAWRLRVYMVYAWGGPLLVAGTGALLDHLPAGAAPLLRPRFAVQRCWFYGDMEILVYFFGPVGVLLLVNLALFISTTRQLTCGLWRRDEVKSTSERAALGRVCAKLVVVMGVTWGADVVSWAAGGPEYVWYATDLLNALQGVLIFLVVGCQPHAWAALKRCAAACCAKGGSGGGERGAGRHSSTHLPSCGESLTHTTQAAAPAPAPVPTPRVPMETVC
Summary
Similarity
Belongs to the G-protein coupled receptor 2 family. Mth subfamily.
Keywords
Cell membrane
Complete proteome
Disulfide bond
G-protein coupled receptor
Glycoprotein
Membrane
Receptor
Reference proteome
Signal
Transducer
Transmembrane
Transmembrane helix
Feature
chain Probable G-protein coupled receptor Mth-like 1
Uniprot
H9J016
A0A212FED2
A0A2H1V6M5
A0A2Z5U769
A0A3S2NQW9
A0A2J7R7M9
+ More
A0A3L8DAZ6 A0A067RCC7 A0A1Z1LVU5 A0A1B6JWB5 A0A0C9R8N0 A0A1J1HUF1 A0A158P3Z3 A0A1B6ENA3 A0A088AVM0 A0A1Q3FUM8 A0A1Q3FUF7 A0A1Q3FUW1 A0A1Q3FUJ4 A0A1Y1LEX7 A0A1Q3FUJ2 A0A1Q3FUS2 A0A2M4AJE5 A0A2M4DM51 A0A2M4DM29 A0A2M3Z501 D6X2H9 A0A0K8SSH1 A0A0A9W856 A0A2M4BHU4 A0A2M4BI25 A0A1B6MAP1 A0A2Y9D3P7 A0A2Y9D0W1 Q7PTM8 A0A0A7P873 A0A146KWL6 A0A1S4G5U6 A0A2M3Z6B7 A0A0A1X380 E0VBJ9 A0A0K8U803 A0A034VEF9 A0A2S2NLW8 A0A0K8VT68 J9K693 W8BGE8 A0A0M4F9I2 A0A2H8TZ60 B4MEK6 B4L1P8 A0A023EU51 B4PXV6 A0A336LR90 A0A336KT42 A0A1A9WWZ5 B4R5V6 A0A1A9X7T3 X2JC86 Q9VXD9 B4IEQ5 A0A1B0G4S0 A0A1B0ALN9 B3NVK0 A0A1A9UUD8 B3MQ68 A0A2S2QEF1 A0A1A9Z4Q7 A0A1I8QE85 B4NQ48 A0A1W4VJ42 A0A1I8MRA8 A0A084VPR6
A0A3L8DAZ6 A0A067RCC7 A0A1Z1LVU5 A0A1B6JWB5 A0A0C9R8N0 A0A1J1HUF1 A0A158P3Z3 A0A1B6ENA3 A0A088AVM0 A0A1Q3FUM8 A0A1Q3FUF7 A0A1Q3FUW1 A0A1Q3FUJ4 A0A1Y1LEX7 A0A1Q3FUJ2 A0A1Q3FUS2 A0A2M4AJE5 A0A2M4DM51 A0A2M4DM29 A0A2M3Z501 D6X2H9 A0A0K8SSH1 A0A0A9W856 A0A2M4BHU4 A0A2M4BI25 A0A1B6MAP1 A0A2Y9D3P7 A0A2Y9D0W1 Q7PTM8 A0A0A7P873 A0A146KWL6 A0A1S4G5U6 A0A2M3Z6B7 A0A0A1X380 E0VBJ9 A0A0K8U803 A0A034VEF9 A0A2S2NLW8 A0A0K8VT68 J9K693 W8BGE8 A0A0M4F9I2 A0A2H8TZ60 B4MEK6 B4L1P8 A0A023EU51 B4PXV6 A0A336LR90 A0A336KT42 A0A1A9WWZ5 B4R5V6 A0A1A9X7T3 X2JC86 Q9VXD9 B4IEQ5 A0A1B0G4S0 A0A1B0ALN9 B3NVK0 A0A1A9UUD8 B3MQ68 A0A2S2QEF1 A0A1A9Z4Q7 A0A1I8QE85 B4NQ48 A0A1W4VJ42 A0A1I8MRA8 A0A084VPR6
Pubmed
19121390
22118469
30249741
24845553
27685537
21347285
+ More
28004739 18362917 19820115 25401762 26823975 12364791 14747013 17210077 25830018 20566863 25348373 24495485 17994087 18057021 24945155 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10908591 25315136 24438588
28004739 18362917 19820115 25401762 26823975 12364791 14747013 17210077 25830018 20566863 25348373 24495485 17994087 18057021 24945155 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10908591 25315136 24438588
EMBL
BABH01018791
AGBW02008947
OWR52116.1
ODYU01000961
SOQ36505.1
AP017499
+ More
BBB06786.1 RSAL01000349 RVE42280.1 NEVH01006727 PNF36848.1 QOIP01000010 RLU17494.1 KK852556 KDR21417.1 KX009474 ARW56807.1 GECU01004247 JAT03460.1 GBYB01012724 JAG82491.1 CVRI01000020 CRK90996.1 ADTU01008628 ADTU01008629 ADTU01008630 ADTU01008631 ADTU01008632 ADTU01008633 ADTU01008634 ADTU01008635 ADTU01008636 ADTU01008637 GECZ01030362 JAS39407.1 GFDL01003807 JAV31238.1 GFDL01003811 JAV31234.1 GFDL01003812 JAV31233.1 GFDL01003810 JAV31235.1 GEZM01062645 JAV69567.1 GFDL01003808 JAV31237.1 GFDL01003809 JAV31236.1 GGFK01007570 MBW40891.1 GGFL01014433 MBW78611.1 GGFL01014434 MBW78612.1 GGFM01002824 MBW23575.1 KQ971372 EFA10672.1 GBRD01009561 JAG56263.1 GBHO01038997 GDHC01019238 JAG04607.1 JAP99390.1 GGFJ01003484 MBW52625.1 GGFJ01003502 MBW52643.1 GEBQ01007053 JAT32924.1 APCN01004112 APCN01004113 AAAB01008797 EAA03645.2 KM588897 AJA06111.1 GDHC01019172 JAP99456.1 GGFM01003311 MBW24062.1 GBXI01009149 GBXI01002869 GBXI01000806 JAD05143.1 JAD11423.1 JAD13486.1 DS235032 EEB10755.1 GDHF01029540 GDHF01011421 JAI22774.1 JAI40893.1 GAKP01018462 GAKP01018461 GAKP01018460 JAC40492.1 GGMR01005157 MBY17776.1 GDHF01026200 GDHF01010240 JAI26114.1 JAI42074.1 ABLF02039939 GAMC01014199 GAMC01014198 GAMC01014197 GAMC01014196 JAB92356.1 CP012528 ALC48882.1 GFXV01007722 MBW19527.1 CH940664 EDW62981.1 CH933810 EDW06701.1 KRF93787.1 GAPW01001151 JAC12447.1 CM000162 EDX01942.2 UFQS01000047 UFQT01000047 SSW98531.1 SSX18917.1 UFQS01000912 UFQT01000912 SSX07619.1 SSX27957.1 CM000366 EDX18106.1 AE014298 AHN59810.1 AY118371 CH480832 EDW46159.1 CCAG010019908 JXJN01000025 JXJN01000026 CH954180 EDV46392.2 CH902621 EDV44494.1 GGMS01006868 MBY76071.1 CH964291 EDW86273.1 ATLV01015029 ATLV01015030 ATLV01015031 ATLV01015032 ATLV01015033 KE524999 KFB39960.1
BBB06786.1 RSAL01000349 RVE42280.1 NEVH01006727 PNF36848.1 QOIP01000010 RLU17494.1 KK852556 KDR21417.1 KX009474 ARW56807.1 GECU01004247 JAT03460.1 GBYB01012724 JAG82491.1 CVRI01000020 CRK90996.1 ADTU01008628 ADTU01008629 ADTU01008630 ADTU01008631 ADTU01008632 ADTU01008633 ADTU01008634 ADTU01008635 ADTU01008636 ADTU01008637 GECZ01030362 JAS39407.1 GFDL01003807 JAV31238.1 GFDL01003811 JAV31234.1 GFDL01003812 JAV31233.1 GFDL01003810 JAV31235.1 GEZM01062645 JAV69567.1 GFDL01003808 JAV31237.1 GFDL01003809 JAV31236.1 GGFK01007570 MBW40891.1 GGFL01014433 MBW78611.1 GGFL01014434 MBW78612.1 GGFM01002824 MBW23575.1 KQ971372 EFA10672.1 GBRD01009561 JAG56263.1 GBHO01038997 GDHC01019238 JAG04607.1 JAP99390.1 GGFJ01003484 MBW52625.1 GGFJ01003502 MBW52643.1 GEBQ01007053 JAT32924.1 APCN01004112 APCN01004113 AAAB01008797 EAA03645.2 KM588897 AJA06111.1 GDHC01019172 JAP99456.1 GGFM01003311 MBW24062.1 GBXI01009149 GBXI01002869 GBXI01000806 JAD05143.1 JAD11423.1 JAD13486.1 DS235032 EEB10755.1 GDHF01029540 GDHF01011421 JAI22774.1 JAI40893.1 GAKP01018462 GAKP01018461 GAKP01018460 JAC40492.1 GGMR01005157 MBY17776.1 GDHF01026200 GDHF01010240 JAI26114.1 JAI42074.1 ABLF02039939 GAMC01014199 GAMC01014198 GAMC01014197 GAMC01014196 JAB92356.1 CP012528 ALC48882.1 GFXV01007722 MBW19527.1 CH940664 EDW62981.1 CH933810 EDW06701.1 KRF93787.1 GAPW01001151 JAC12447.1 CM000162 EDX01942.2 UFQS01000047 UFQT01000047 SSW98531.1 SSX18917.1 UFQS01000912 UFQT01000912 SSX07619.1 SSX27957.1 CM000366 EDX18106.1 AE014298 AHN59810.1 AY118371 CH480832 EDW46159.1 CCAG010019908 JXJN01000025 JXJN01000026 CH954180 EDV46392.2 CH902621 EDV44494.1 GGMS01006868 MBY76071.1 CH964291 EDW86273.1 ATLV01015029 ATLV01015030 ATLV01015031 ATLV01015032 ATLV01015033 KE524999 KFB39960.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000235965
UP000279307
UP000027135
+ More
UP000183832 UP000005205 UP000005203 UP000007266 UP000076407 UP000075840 UP000007062 UP000009046 UP000007819 UP000092553 UP000008792 UP000009192 UP000002282 UP000091820 UP000000304 UP000092443 UP000000803 UP000001292 UP000092444 UP000092460 UP000008711 UP000078200 UP000007801 UP000092445 UP000095300 UP000007798 UP000192221 UP000095301 UP000030765
UP000183832 UP000005205 UP000005203 UP000007266 UP000076407 UP000075840 UP000007062 UP000009046 UP000007819 UP000092553 UP000008792 UP000009192 UP000002282 UP000091820 UP000000304 UP000092443 UP000000803 UP000001292 UP000092444 UP000092460 UP000008711 UP000078200 UP000007801 UP000092445 UP000095300 UP000007798 UP000192221 UP000095301 UP000030765
Interpro
SUPFAM
SSF63877
SSF63877
ProteinModelPortal
H9J016
A0A212FED2
A0A2H1V6M5
A0A2Z5U769
A0A3S2NQW9
A0A2J7R7M9
+ More
A0A3L8DAZ6 A0A067RCC7 A0A1Z1LVU5 A0A1B6JWB5 A0A0C9R8N0 A0A1J1HUF1 A0A158P3Z3 A0A1B6ENA3 A0A088AVM0 A0A1Q3FUM8 A0A1Q3FUF7 A0A1Q3FUW1 A0A1Q3FUJ4 A0A1Y1LEX7 A0A1Q3FUJ2 A0A1Q3FUS2 A0A2M4AJE5 A0A2M4DM51 A0A2M4DM29 A0A2M3Z501 D6X2H9 A0A0K8SSH1 A0A0A9W856 A0A2M4BHU4 A0A2M4BI25 A0A1B6MAP1 A0A2Y9D3P7 A0A2Y9D0W1 Q7PTM8 A0A0A7P873 A0A146KWL6 A0A1S4G5U6 A0A2M3Z6B7 A0A0A1X380 E0VBJ9 A0A0K8U803 A0A034VEF9 A0A2S2NLW8 A0A0K8VT68 J9K693 W8BGE8 A0A0M4F9I2 A0A2H8TZ60 B4MEK6 B4L1P8 A0A023EU51 B4PXV6 A0A336LR90 A0A336KT42 A0A1A9WWZ5 B4R5V6 A0A1A9X7T3 X2JC86 Q9VXD9 B4IEQ5 A0A1B0G4S0 A0A1B0ALN9 B3NVK0 A0A1A9UUD8 B3MQ68 A0A2S2QEF1 A0A1A9Z4Q7 A0A1I8QE85 B4NQ48 A0A1W4VJ42 A0A1I8MRA8 A0A084VPR6
A0A3L8DAZ6 A0A067RCC7 A0A1Z1LVU5 A0A1B6JWB5 A0A0C9R8N0 A0A1J1HUF1 A0A158P3Z3 A0A1B6ENA3 A0A088AVM0 A0A1Q3FUM8 A0A1Q3FUF7 A0A1Q3FUW1 A0A1Q3FUJ4 A0A1Y1LEX7 A0A1Q3FUJ2 A0A1Q3FUS2 A0A2M4AJE5 A0A2M4DM51 A0A2M4DM29 A0A2M3Z501 D6X2H9 A0A0K8SSH1 A0A0A9W856 A0A2M4BHU4 A0A2M4BI25 A0A1B6MAP1 A0A2Y9D3P7 A0A2Y9D0W1 Q7PTM8 A0A0A7P873 A0A146KWL6 A0A1S4G5U6 A0A2M3Z6B7 A0A0A1X380 E0VBJ9 A0A0K8U803 A0A034VEF9 A0A2S2NLW8 A0A0K8VT68 J9K693 W8BGE8 A0A0M4F9I2 A0A2H8TZ60 B4MEK6 B4L1P8 A0A023EU51 B4PXV6 A0A336LR90 A0A336KT42 A0A1A9WWZ5 B4R5V6 A0A1A9X7T3 X2JC86 Q9VXD9 B4IEQ5 A0A1B0G4S0 A0A1B0ALN9 B3NVK0 A0A1A9UUD8 B3MQ68 A0A2S2QEF1 A0A1A9Z4Q7 A0A1I8QE85 B4NQ48 A0A1W4VJ42 A0A1I8MRA8 A0A084VPR6
PDB
5UZ7
E-value=0.0120071,
Score=93
Ontologies
GO
Topology
Subcellular location
Cell membrane
SignalP
Position: 1 - 23,
Likelihood: 0.936439
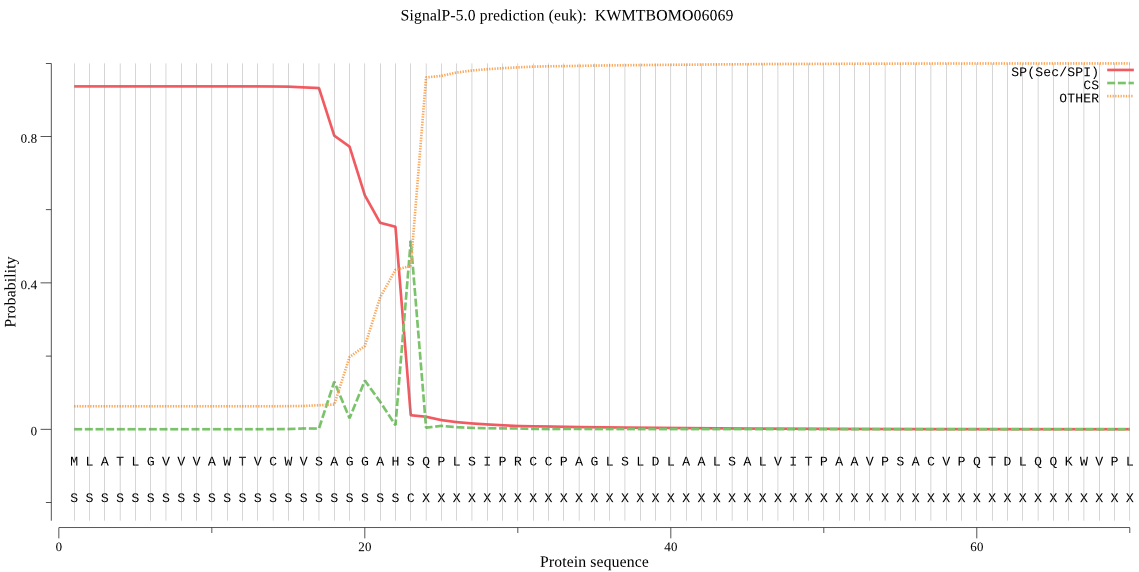
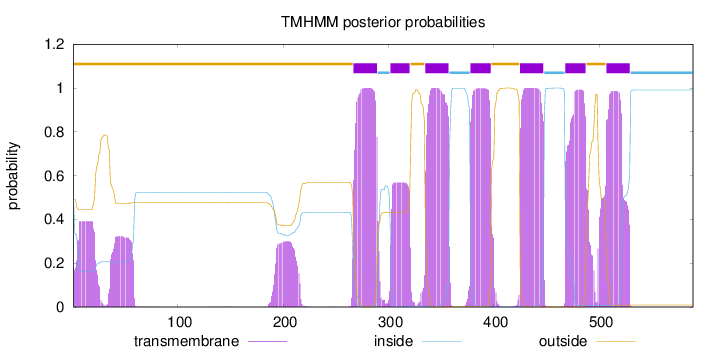
Length:
589
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
163.7515
Exp number, first 60 AAs:
14.76628
Total prob of N-in:
0.50984
POSSIBLE N-term signal
sequence
outside
1 - 266
TMhelix
267 - 289
inside
290 - 301
TMhelix
302 - 320
outside
321 - 334
TMhelix
335 - 357
inside
358 - 377
TMhelix
378 - 397
outside
398 - 424
TMhelix
425 - 447
inside
448 - 467
TMhelix
468 - 487
outside
488 - 506
TMhelix
507 - 529
inside
530 - 589
Population Genetic Test Statistics
Pi
2.277868
Theta
3.951555
Tajima's D
-1.295233
CLR
11.859473
CSRT
0.0824458777061147
Interpretation
Uncertain
