Gene
KWMTBOMO06058 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002847
Annotation
PREDICTED:_uncharacterized_protein_LOC106713873_isoform_X1_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 2.953
Sequence
CDS
ATGGCGTCGCCAAGATTTTCAACCATTTTGCTAGCAGCATTCCATCTTTTTACCGTATGTTGGGGAGGTCCGGTGGGTCCGCTACCGAATGTAACGAGCGTCGATTTGGGACTTAAAGAAGGTAGTTGCGCGCTAGGAGACGTGGTATATCTTCCCGGAGACGAATTCCCAGGTTCAGACCCCTGCGAGAAGTGCAAGTGCTCCGATGGAGGGGGGGTCCAATGTGAGAAGCAGCAATGCGAATCCAGGCCTGGCTGCAAGGCTGTGCATCGACCCGACCATTGCTGTCCAACGTATCAGTGTGATTGCGAGCAAGAAGGTCGTGTCTACGATAACGGTGAAAAGTTGGTCGATCCTGCAGATCCCTGCCGCGTGTGCTACTGCCAGGGGGGAGAGGTAGTCTGCAGACGGATCGCTTGCTTCGTCAGAGATGACTGCACTCCTCGGCTTGTGCCCGGTAGATGTTGTCCTGAATACGACAACTGTCCTGTCAGAGGGGTAACCATGCTTCCTGGCGTCTCTTCCTTGCCGCCCAACCTTTCACCTACAGAAGATATCAAGGGTAGTGCTGCACCAACTGCTCCTAAGGAAAATCTTAAACAAGAATTCACTATTAAGGAAATAACACCAGTATCTGAGATTCCTATAATGAATGATGTGAAAATTAAAAAGATCTTACCATTTCCAAGTATCGACGTTGCCGAGTATTCATCGTCGAAGTCTCCTCCTATCACAAGAGAAGCTACTTCTGAGATATCCGTTAAAACAGAATCTGAAATCGTCAAAACGGACGCTCCTACGATCACTGAAGTCCATCAGAAAGAGAGTGACAAGAAGCCAGATGATACACAGCCCTCTAAAATAAGTTTTTCTACTCAAGATTCTATTAACAGTGATATTTATTCGTCGAATGAATCAACTGTCACAAATGGATTACCTACAGCTCCATCTGTGACTACAACTAAAGCACCGATCATTGAAGAAGAAGACACATCACTTTTTGATCATAACCCAGCATTCCCACCTATTCCAGACGATTTAGCAGTCTTGAGCAACCACGGAGATGAAATCGTACCCGAGCCATCTGCAGAGGGCGACCACATGACTGTGCACGACACCGTTCCCAGCTTGCCTTCAGTAGAGCCAAAGCTTGAAGAACCTATTGAACCTAAAGAAGTCACTGACATTATACTGACAACTGAAAAACCATCGGTCACTTCAATCGAAACTAGTACAACAGCTAAACAAGAAAGCACTACAGTTGTAGAACAGCTCAAAGAAAGCCCAATGCTTAATTTGAGGTCCGCTATACCAACTGAGATACTCAACGCTCCATCTCTTGTACCAGCAGACACCGGGGAGCCGTTAGATGATGGCATAACTACTGATGTACCAGAAATATCCTCAACCACAGAGGAAACCGAAACGTCCAAAGAAAGCTCTAACCACACTATTGAAGTTTCTGAAACTCCTGTTATCGAATCTTTTGCTACAGCTACTGGTGCAGAATTAGACACAACCACTCCTAAAGTATCCACTGAACATGAAATTGAAGTCACCACTAAAGACCTGGAACAAACGACCAGTTCGATAGCAAAATCAAATAGCGCAACGGAAGTGACGTCCCATACTGAAATTAGTTCTTTACCGATTGAAACGAGTGATCAAAATCCACACGAGTCTGAAGCTGTGGTGAAAGATAAAATCGTAGACTCTACTATCGACCCTTTTGCAACTAGCGCTAAGGTTTCACCGTCTGAAGTAATAACCGTTTCAAGGGCGGTGTCAAATGAAGATAACGTATTTGAAAACGTTGAAACTACTGAATTTATTTTAACATCGTTTGGCTCTTCCGAAACTGCCACCGATTCTGTCGAATTGATCAAAATATCTGGTGATCCTGAGAAAAGTGCTCAGATAGTTGTAGAGCCTGAAGAGAAGAGGAATAGTGTACTCGCTGATTTAATTAATCTAGTTAGCGACGTAGCCACGATTAACGACCACACTGAAAACCCTAATAAGGAACAAGCACCGAAACCAACTAGTATATCTGATTCTGAAGAACTCATACCAGTCAACGCTGGGTACAAAAGCAAGAACAGTAATTATAATCAAAACTCTATAACTGAAATTCCTTACAAATCAAAAAATACTCCAGGAAACCGTCAAAAAGTAGTCGAAATAGAAGACGACGAATCTGAGAGTATTACTGATCTTCCGGCACCACACGACAAGGTGGAACCTACCACACGCCGACCGATTATCGATAATGTGTCGGACAAAAAGATGGAGAACAAAACTCAGGAAAAAGATACCGAAATCATAACGCAGTCTTACGTCCCGACGATCAACAGACGGCCGATAAAGGTCATCATGAAGAAAAATAACGACAGGCCGGTTACTGAGTCACCCGAAGGCTCACTGGTATCTGTCTCGAGTGAGGCCTTTACTGATGACATAACCGACACCACAGAGAATAGCGTGGCTGCAGAGGAGAAGAAACGTACAGAAGATTCCACGGAAACCGTGCTGGTCACAGAGGCTCAATGA
Protein
MASPRFSTILLAAFHLFTVCWGGPVGPLPNVTSVDLGLKEGSCALGDVVYLPGDEFPGSDPCEKCKCSDGGGVQCEKQQCESRPGCKAVHRPDHCCPTYQCDCEQEGRVYDNGEKLVDPADPCRVCYCQGGEVVCRRIACFVRDDCTPRLVPGRCCPEYDNCPVRGVTMLPGVSSLPPNLSPTEDIKGSAAPTAPKENLKQEFTIKEITPVSEIPIMNDVKIKKILPFPSIDVAEYSSSKSPPITREATSEISVKTESEIVKTDAPTITEVHQKESDKKPDDTQPSKISFSTQDSINSDIYSSNESTVTNGLPTAPSVTTTKAPIIEEEDTSLFDHNPAFPPIPDDLAVLSNHGDEIVPEPSAEGDHMTVHDTVPSLPSVEPKLEEPIEPKEVTDIILTTEKPSVTSIETSTTAKQESTTVVEQLKESPMLNLRSAIPTEILNAPSLVPADTGEPLDDGITTDVPEISSTTEETETSKESSNHTIEVSETPVIESFATATGAELDTTTPKVSTEHEIEVTTKDLEQTTSSIAKSNSATEVTSHTEISSLPIETSDQNPHESEAVVKDKIVDSTIDPFATSAKVSPSEVITVSRAVSNEDNVFENVETTEFILTSFGSSETATDSVELIKISGDPEKSAQIVVEPEEKRNSVLADLINLVSDVATINDHTENPNKEQAPKPTSISDSEELIPVNAGYKSKNSNYNQNSITEIPYKSKNTPGNRQKVVEIEDDESESITDLPAPHDKVEPTTRRPIIDNVSDKKMENKTQEKDTEIITQSYVPTINRRPIKVIMKKNNDRPVTESPEGSLVSVSSEAFTDDITDTTENSVAAEEKKRTEDSTETVLVTEAQ
Summary
Uniprot
EMBL
Proteomes
PRIDE
Interpro
IPR001007
VWF_dom
ProteinModelPortal
PDB
1U5M
E-value=0.00332702,
Score=99
Ontologies
GO
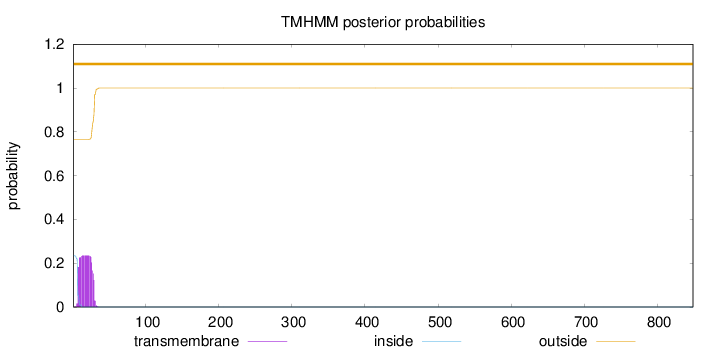
Topology
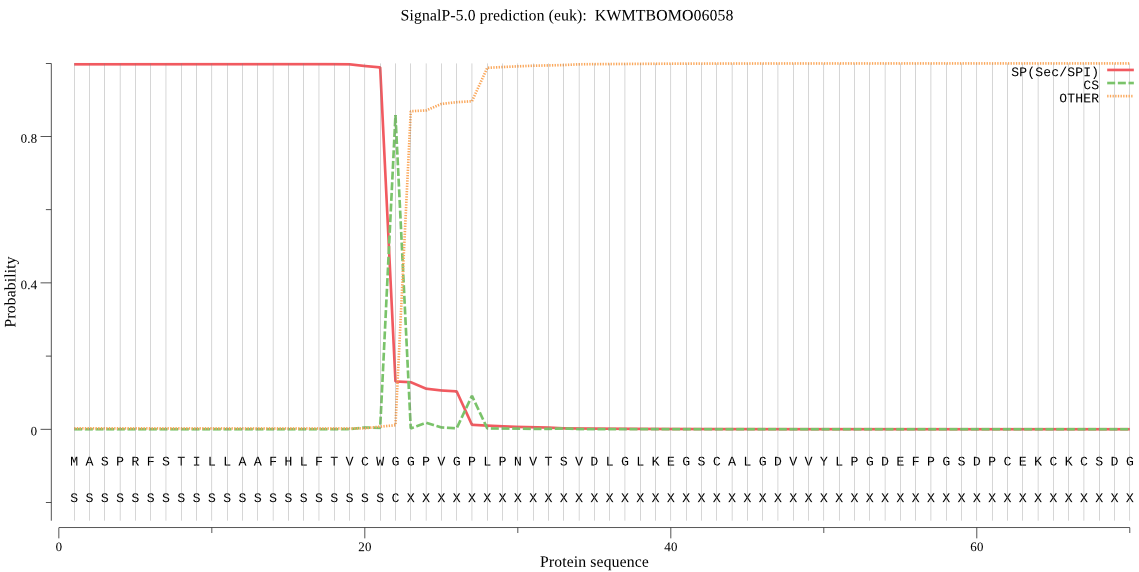
SignalP
Position: 1 - 22,
Likelihood: 0.997730
Length:
849
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.07619
Exp number, first 60 AAs:
5.07619
Total prob of N-in:
0.23532
outside
1 - 849
Population Genetic Test Statistics
Pi
27.463111
Theta
23.562917
Tajima's D
-1.386718
CLR
0.684866
CSRT
0.0742462876856157
Interpretation
Uncertain
