Gene
KWMTBOMO06055
Pre Gene Modal
BGIBMGA002845
Annotation
PREDICTED:_vacuolar_protein_sorting-associated_protein_13D-like_[Plutella_xylostella]
Full name
Vacuolar protein sorting-associated protein 13D
Location in the cell
Mitochondrial Reliability : 1.839
Sequence
CDS
ATGACAAAGGGGGGTATGACGTCACTGGTGAAACACAGCTACGAAGGCGCGACGCAGGAGGGGCTGGGAGGGTTCCTGACGGGGGTGGGGAAGGGGCTGGTGGGGACCGTCACCAAGCCTGTCATCGGCGTCCTCGACCTCGCTGCTGAGACGGCTTCGGCACTCAGGGACACCAGTCGAAGGTCGGACAAGTGGGTGCCGGCGCGGGTGCGCGGGCCGCGGTGCGCGTGGGGCGCGGGCTCCGCCCTGCCGCGCTACTGCGGGGCGCAGGCCCACGGCGCGGCGCTGCTGTACGCGCTCAACAACAACGACTACTCCGAGCGCTTCCTGGCCTACCGCATCGTCCGGGACACGCCGCACGACATCCGCGCGCTGCTCTCGGACTCCTACCTGCGCATCTTCACCTGCAAGCACTCTGCGCCGCAGGTCGTCATGGAGACGCACCTCAGCAACCTGGTGTCGTGCGCGGTGGTGGAGGCCGGCGGCGCGCAGCTGGTGGAGCTGGGCGTGCGCGGCGGCGAGGCGGGCGTGCGGCGGCCGCGCGTGCAGTGCGACTCGGCGGCGCTGGCGCGCTGGCTGCGCACGCACGCGCTGTACGCCGCGCAGCTCTACCACGAGCGGGCGCACACGCTGCTGCCGCCCGAGCCCGCACTCTGCGCGCCCGGGAACCATTAA
Protein
MTKGGMTSLVKHSYEGATQEGLGGFLTGVGKGLVGTVTKPVIGVLDLAAETASALRDTSRRSDKWVPARVRGPRCAWGAGSALPRYCGAQAHGAALLYALNNNDYSERFLAYRIVRDTPHDIRALLSDSYLRIFTCKHSAPQVVMETHLSNLVSCAVVEAGGAQLVELGVRGGEAGVRRPRVQCDSAALARWLRTHALYAAQLYHERAHTLLPPEPALCAPGNH
Summary
Description
Functions in promoting mitochondrial clearance by mitochondrial autophagy (mitophagy), also possibly by positively regulating mitochondrial fission (PubMed:29307555). Mitophagy plays an important role in regulating cell health and mitochondrial size and homeostasis (PubMed:29307555).
Similarity
Belongs to the VPS13 family.
Keywords
Complete proteome
Cytoplasm
Lectin
Lysosome
Reference proteome
Feature
chain Vacuolar protein sorting-associated protein 13D
Uniprot
A0A2H1V893
A0A2W1BLQ7
A0A2A4J3I8
A0A0L7L9Q2
A0A194Q3V3
A0A212FED5
+ More
A0A2J7RLT2 A0A3S2LUG1 A0A2J7RLT0 A0A2J7RLV8 A0A1B6C7B3 A0A1Y1L7W2 A0A1B6D961 A0A1B6CRD8 A0A146KXH9 A0A0A9W7Q2 A0A0A9WG36 D6X361 N6TGW4 E0VE59 E9FVT2 A0A1B6CA12 A0A146MF30 T1I7N4 A0A232FP36 K7ITC6 A0A067QRL8 B4HG59 B4QRR2 A0A0J9UK05 A0A1B6MEQ3 B3M9D0 A0A0R1DXM4 Q9VU08 B4PGU5 E2BY65 A0A0L7QSM4 A0A2A3EG84 A0A1B6FCY4 A0A0M9A200 A0A1B6J1C2 A0A0P5M4A7 A0A0P5L7A1 A0A0P5KC92 A0A0P5KVU5 A0A0P6EQK7 A0A0P5L303 A0A0P5YV10 A0A0N8BKM7 A0A0P5PGL9 A0A0P5M5H5 A0A0N8AWY6 A0A0P5FY43 A0A0P5KFU5 A0A0P6EXV1 A0A0P5LEA2 A0A0P5LP71 A0A0P6FQM8 A0A0P6EDP8 A0A0P6GRV7 A0A0P5GFK9 A0A0P5NNC8 A0A0P5NDQ5 A0A0N8BUD2 A0A0P5NK87 A0A0N8ARY6 A0A0P5FY99 A0A0P5JRV7 A0A0N8EIS0 A0A0P5JJQ0 A0A0N8B217 A0A0P5I0D6 A0A0P5KIX8 A0A0P5L047 A0A0P6BL35 A0A0P6IBF0 A0A0P5PPU7 A0A0P5H3J7 A0A0N8E9P1 A0A0P5LFX4 A0A0P6FA87 A0A0P5RJZ5 A0A0P5JN25 A0A0P5NND2 A0A0P5QFL0 A0A0P6GTE3 A0A0P6E410 A0A0N8C0A0 A0A0P5MJQ0 A0A0P6D444 A0A0P5R6D8 A0A0P5S1F0 A0A0P5RSH3 A0A0P5HI48 A0A0N8BMH0 A0A0P5M8U8 A0A0P5P060 A0A0P5NH29
A0A2J7RLT2 A0A3S2LUG1 A0A2J7RLT0 A0A2J7RLV8 A0A1B6C7B3 A0A1Y1L7W2 A0A1B6D961 A0A1B6CRD8 A0A146KXH9 A0A0A9W7Q2 A0A0A9WG36 D6X361 N6TGW4 E0VE59 E9FVT2 A0A1B6CA12 A0A146MF30 T1I7N4 A0A232FP36 K7ITC6 A0A067QRL8 B4HG59 B4QRR2 A0A0J9UK05 A0A1B6MEQ3 B3M9D0 A0A0R1DXM4 Q9VU08 B4PGU5 E2BY65 A0A0L7QSM4 A0A2A3EG84 A0A1B6FCY4 A0A0M9A200 A0A1B6J1C2 A0A0P5M4A7 A0A0P5L7A1 A0A0P5KC92 A0A0P5KVU5 A0A0P6EQK7 A0A0P5L303 A0A0P5YV10 A0A0N8BKM7 A0A0P5PGL9 A0A0P5M5H5 A0A0N8AWY6 A0A0P5FY43 A0A0P5KFU5 A0A0P6EXV1 A0A0P5LEA2 A0A0P5LP71 A0A0P6FQM8 A0A0P6EDP8 A0A0P6GRV7 A0A0P5GFK9 A0A0P5NNC8 A0A0P5NDQ5 A0A0N8BUD2 A0A0P5NK87 A0A0N8ARY6 A0A0P5FY99 A0A0P5JRV7 A0A0N8EIS0 A0A0P5JJQ0 A0A0N8B217 A0A0P5I0D6 A0A0P5KIX8 A0A0P5L047 A0A0P6BL35 A0A0P6IBF0 A0A0P5PPU7 A0A0P5H3J7 A0A0N8E9P1 A0A0P5LFX4 A0A0P6FA87 A0A0P5RJZ5 A0A0P5JN25 A0A0P5NND2 A0A0P5QFL0 A0A0P6GTE3 A0A0P6E410 A0A0N8C0A0 A0A0P5MJQ0 A0A0P6D444 A0A0P5R6D8 A0A0P5S1F0 A0A0P5RSH3 A0A0P5HI48 A0A0N8BMH0 A0A0P5M8U8 A0A0P5P060 A0A0P5NH29
Pubmed
EMBL
ODYU01001174
SOQ37029.1
KZ150039
PZC74595.1
NWSH01003638
PCG66010.1
+ More
JTDY01002066 KOB72192.1 KQ459472 KPJ00217.1 AGBW02008947 OWR52102.1 NEVH01002683 PNF41792.1 RSAL01000192 RVE44644.1 PNF41791.1 PNF41789.1 GEDC01027911 JAS09387.1 GEZM01066271 JAV68025.1 GEDC01015106 GEDC01006474 JAS22192.1 JAS30824.1 GEDC01021320 JAS15978.1 GDHC01017701 JAQ00928.1 GBHO01039780 JAG03824.1 GBHO01039779 JAG03825.1 KQ971372 EFA09797.2 APGK01038542 KB740960 ENN77018.1 DS235088 EEB11665.1 GL732525 EFX89051.1 GEDC01026972 JAS10326.1 GDHC01001303 JAQ17326.1 ACPB03000178 ACPB03000179 ACPB03000180 NNAY01000006 OXU32127.1 AAZX01003330 KK853018 KDR12416.1 CH480815 EDW41305.1 CM000363 EDX10276.1 CM002912 KMY99285.1 GEBQ01005612 JAT34365.1 CH902618 EDV41143.2 CM000159 KRK01769.1 AE014296 EDW94334.2 GL451420 EFN79344.1 KQ414756 KOC61642.1 KZ288266 PBC30166.1 GECZ01021709 JAS48060.1 KQ435760 KOX75567.1 GECU01014743 JAS92963.1 GDIQ01172388 JAK79337.1 GDIQ01175636 JAK76089.1 GDIQ01186336 JAK65389.1 GDIQ01186337 GDIQ01186334 JAK65391.1 GDIQ01059233 JAN35504.1 GDIQ01175638 JAK76087.1 GDIP01053078 JAM50637.1 GDIQ01168443 JAK83282.1 GDIQ01128760 JAL22966.1 GDIQ01163297 JAK88428.1 GDIQ01235921 GDIQ01234771 JAK16954.1 GDIQ01248768 JAK02957.1 GDIQ01186335 JAK65390.1 GDIQ01056585 JAN38152.1 GDIQ01172390 JAK79335.1 GDIQ01175637 JAK76088.1 GDIQ01044641 JAN50096.1 GDIQ01078422 JAN16315.1 GDIQ01031045 JAN63692.1 GDIQ01248770 JAK02955.1 GDIQ01144005 JAL07721.1 GDIQ01144004 JAL07722.1 GDIQ01144003 JAL07723.1 GDIQ01144006 GDIQ01144002 JAL07720.1 GDIQ01248771 JAK02954.1 GDIQ01248774 GDIQ01248773 GDIQ01248772 GDIQ01248769 JAK02953.1 GDIQ01220524 JAK31201.1 GDIQ01022708 JAN72029.1 GDIQ01201928 JAK49797.1 GDIQ01220523 JAK31202.1 GDIQ01220527 GDIQ01220526 GDIQ01220525 JAK31200.1 GDIQ01184687 JAK67038.1 GDIQ01184688 GDIQ01184686 JAK67037.1 GDIP01016461 JAM87254.1 GDIQ01006963 JAN87774.1 GDIQ01136008 JAL15718.1 GDIQ01235872 JAK15853.1 GDIQ01048140 JAN46597.1 GDIQ01170143 JAK81582.1 GDIQ01063336 JAN31401.1 GDIQ01099668 JAL52058.1 GDIQ01200730 JAK50995.1 GDIQ01146754 JAL04972.1 GDIQ01136007 JAL15719.1 GDIQ01049639 JAN45098.1 GDIQ01080322 JAN14415.1 GDIQ01127459 JAL24267.1 GDIQ01162745 JAK88980.1 GDIQ01083023 JAN11714.1 GDIQ01112060 JAL39666.1 GDIQ01098316 JAL53410.1 GDIQ01105965 JAL45761.1 GDIQ01229271 JAK22454.1 GDIQ01163299 JAK88426.1 GDIQ01163296 JAK88429.1 GDIQ01145407 JAL06319.1 GDIQ01145405 JAL06321.1
JTDY01002066 KOB72192.1 KQ459472 KPJ00217.1 AGBW02008947 OWR52102.1 NEVH01002683 PNF41792.1 RSAL01000192 RVE44644.1 PNF41791.1 PNF41789.1 GEDC01027911 JAS09387.1 GEZM01066271 JAV68025.1 GEDC01015106 GEDC01006474 JAS22192.1 JAS30824.1 GEDC01021320 JAS15978.1 GDHC01017701 JAQ00928.1 GBHO01039780 JAG03824.1 GBHO01039779 JAG03825.1 KQ971372 EFA09797.2 APGK01038542 KB740960 ENN77018.1 DS235088 EEB11665.1 GL732525 EFX89051.1 GEDC01026972 JAS10326.1 GDHC01001303 JAQ17326.1 ACPB03000178 ACPB03000179 ACPB03000180 NNAY01000006 OXU32127.1 AAZX01003330 KK853018 KDR12416.1 CH480815 EDW41305.1 CM000363 EDX10276.1 CM002912 KMY99285.1 GEBQ01005612 JAT34365.1 CH902618 EDV41143.2 CM000159 KRK01769.1 AE014296 EDW94334.2 GL451420 EFN79344.1 KQ414756 KOC61642.1 KZ288266 PBC30166.1 GECZ01021709 JAS48060.1 KQ435760 KOX75567.1 GECU01014743 JAS92963.1 GDIQ01172388 JAK79337.1 GDIQ01175636 JAK76089.1 GDIQ01186336 JAK65389.1 GDIQ01186337 GDIQ01186334 JAK65391.1 GDIQ01059233 JAN35504.1 GDIQ01175638 JAK76087.1 GDIP01053078 JAM50637.1 GDIQ01168443 JAK83282.1 GDIQ01128760 JAL22966.1 GDIQ01163297 JAK88428.1 GDIQ01235921 GDIQ01234771 JAK16954.1 GDIQ01248768 JAK02957.1 GDIQ01186335 JAK65390.1 GDIQ01056585 JAN38152.1 GDIQ01172390 JAK79335.1 GDIQ01175637 JAK76088.1 GDIQ01044641 JAN50096.1 GDIQ01078422 JAN16315.1 GDIQ01031045 JAN63692.1 GDIQ01248770 JAK02955.1 GDIQ01144005 JAL07721.1 GDIQ01144004 JAL07722.1 GDIQ01144003 JAL07723.1 GDIQ01144006 GDIQ01144002 JAL07720.1 GDIQ01248771 JAK02954.1 GDIQ01248774 GDIQ01248773 GDIQ01248772 GDIQ01248769 JAK02953.1 GDIQ01220524 JAK31201.1 GDIQ01022708 JAN72029.1 GDIQ01201928 JAK49797.1 GDIQ01220523 JAK31202.1 GDIQ01220527 GDIQ01220526 GDIQ01220525 JAK31200.1 GDIQ01184687 JAK67038.1 GDIQ01184688 GDIQ01184686 JAK67037.1 GDIP01016461 JAM87254.1 GDIQ01006963 JAN87774.1 GDIQ01136008 JAL15718.1 GDIQ01235872 JAK15853.1 GDIQ01048140 JAN46597.1 GDIQ01170143 JAK81582.1 GDIQ01063336 JAN31401.1 GDIQ01099668 JAL52058.1 GDIQ01200730 JAK50995.1 GDIQ01146754 JAL04972.1 GDIQ01136007 JAL15719.1 GDIQ01049639 JAN45098.1 GDIQ01080322 JAN14415.1 GDIQ01127459 JAL24267.1 GDIQ01162745 JAK88980.1 GDIQ01083023 JAN11714.1 GDIQ01112060 JAL39666.1 GDIQ01098316 JAL53410.1 GDIQ01105965 JAL45761.1 GDIQ01229271 JAK22454.1 GDIQ01163299 JAK88426.1 GDIQ01163296 JAK88429.1 GDIQ01145407 JAL06319.1 GDIQ01145405 JAL06321.1
Proteomes
PRIDE
Pfam
Interpro
ProteinModelPortal
A0A2H1V893
A0A2W1BLQ7
A0A2A4J3I8
A0A0L7L9Q2
A0A194Q3V3
A0A212FED5
+ More
A0A2J7RLT2 A0A3S2LUG1 A0A2J7RLT0 A0A2J7RLV8 A0A1B6C7B3 A0A1Y1L7W2 A0A1B6D961 A0A1B6CRD8 A0A146KXH9 A0A0A9W7Q2 A0A0A9WG36 D6X361 N6TGW4 E0VE59 E9FVT2 A0A1B6CA12 A0A146MF30 T1I7N4 A0A232FP36 K7ITC6 A0A067QRL8 B4HG59 B4QRR2 A0A0J9UK05 A0A1B6MEQ3 B3M9D0 A0A0R1DXM4 Q9VU08 B4PGU5 E2BY65 A0A0L7QSM4 A0A2A3EG84 A0A1B6FCY4 A0A0M9A200 A0A1B6J1C2 A0A0P5M4A7 A0A0P5L7A1 A0A0P5KC92 A0A0P5KVU5 A0A0P6EQK7 A0A0P5L303 A0A0P5YV10 A0A0N8BKM7 A0A0P5PGL9 A0A0P5M5H5 A0A0N8AWY6 A0A0P5FY43 A0A0P5KFU5 A0A0P6EXV1 A0A0P5LEA2 A0A0P5LP71 A0A0P6FQM8 A0A0P6EDP8 A0A0P6GRV7 A0A0P5GFK9 A0A0P5NNC8 A0A0P5NDQ5 A0A0N8BUD2 A0A0P5NK87 A0A0N8ARY6 A0A0P5FY99 A0A0P5JRV7 A0A0N8EIS0 A0A0P5JJQ0 A0A0N8B217 A0A0P5I0D6 A0A0P5KIX8 A0A0P5L047 A0A0P6BL35 A0A0P6IBF0 A0A0P5PPU7 A0A0P5H3J7 A0A0N8E9P1 A0A0P5LFX4 A0A0P6FA87 A0A0P5RJZ5 A0A0P5JN25 A0A0P5NND2 A0A0P5QFL0 A0A0P6GTE3 A0A0P6E410 A0A0N8C0A0 A0A0P5MJQ0 A0A0P6D444 A0A0P5R6D8 A0A0P5S1F0 A0A0P5RSH3 A0A0P5HI48 A0A0N8BMH0 A0A0P5M8U8 A0A0P5P060 A0A0P5NH29
A0A2J7RLT2 A0A3S2LUG1 A0A2J7RLT0 A0A2J7RLV8 A0A1B6C7B3 A0A1Y1L7W2 A0A1B6D961 A0A1B6CRD8 A0A146KXH9 A0A0A9W7Q2 A0A0A9WG36 D6X361 N6TGW4 E0VE59 E9FVT2 A0A1B6CA12 A0A146MF30 T1I7N4 A0A232FP36 K7ITC6 A0A067QRL8 B4HG59 B4QRR2 A0A0J9UK05 A0A1B6MEQ3 B3M9D0 A0A0R1DXM4 Q9VU08 B4PGU5 E2BY65 A0A0L7QSM4 A0A2A3EG84 A0A1B6FCY4 A0A0M9A200 A0A1B6J1C2 A0A0P5M4A7 A0A0P5L7A1 A0A0P5KC92 A0A0P5KVU5 A0A0P6EQK7 A0A0P5L303 A0A0P5YV10 A0A0N8BKM7 A0A0P5PGL9 A0A0P5M5H5 A0A0N8AWY6 A0A0P5FY43 A0A0P5KFU5 A0A0P6EXV1 A0A0P5LEA2 A0A0P5LP71 A0A0P6FQM8 A0A0P6EDP8 A0A0P6GRV7 A0A0P5GFK9 A0A0P5NNC8 A0A0P5NDQ5 A0A0N8BUD2 A0A0P5NK87 A0A0N8ARY6 A0A0P5FY99 A0A0P5JRV7 A0A0N8EIS0 A0A0P5JJQ0 A0A0N8B217 A0A0P5I0D6 A0A0P5KIX8 A0A0P5L047 A0A0P6BL35 A0A0P6IBF0 A0A0P5PPU7 A0A0P5H3J7 A0A0N8E9P1 A0A0P5LFX4 A0A0P6FA87 A0A0P5RJZ5 A0A0P5JN25 A0A0P5NND2 A0A0P5QFL0 A0A0P6GTE3 A0A0P6E410 A0A0N8C0A0 A0A0P5MJQ0 A0A0P6D444 A0A0P5R6D8 A0A0P5S1F0 A0A0P5RSH3 A0A0P5HI48 A0A0N8BMH0 A0A0P5M8U8 A0A0P5P060 A0A0P5NH29
Ontologies
GO
PANTHER
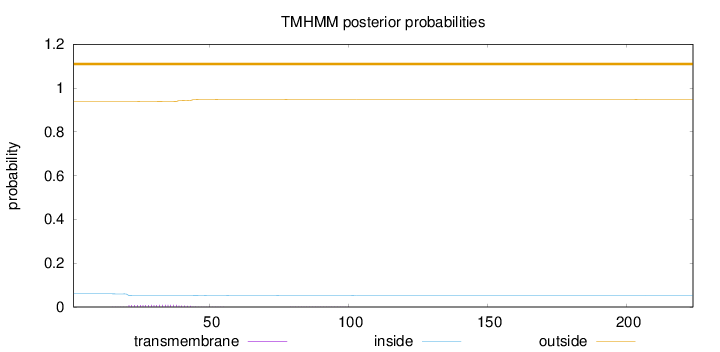
Topology
Subcellular location
Cytoplasm
Lysosome
Lysosome
Length:
224
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.21666
Exp number, first 60 AAs:
0.20863
Total prob of N-in:
0.06003
outside
1 - 224
Population Genetic Test Statistics
Pi
14.066319
Theta
14.173352
Tajima's D
-0.575913
CLR
1.017599
CSRT
0.217639118044098
Interpretation
Uncertain
