Gene
KWMTBOMO06048
Pre Gene Modal
BGIBMGA002841
Annotation
caspase_Nc_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.791
Sequence
CDS
ATGACAAAATTTTACATATACTTAAGACCGAGGCAGGGCTCGGACGCGCCCGTGCCGCCGCCGCCCGTGAACAGCAGTGAGCCGGTCGCCATCCCGTGCTTCCACGTGAAGAAGAGCACTCACTTCTTCGAGGACGATGTGAGCAGGGATATCCAGTTGTACCGCACGCGGAGCCGGAAGCGAGGAGTTCTAGTCGTGTTCAGCTACATTTCGTTCCTGCTGAACATCGAGCCCAACAGGGACGGGGTCAATGTGGACTGCGACAAGTTGAAGTACTTATTCTCAGAGATCGGCTTCGAGGTGCTCACTTACCAGAACTTGACTTTGGAGCAAACCCGCGCGACCCTGGAGCAGATGTCGTCGGTGCTGGCGGGCTACGAGTGCGTGTTCCTGGTGGTGTCGTCGCACGGCCACGAGCGCCGCTACTCCTCCGACACGGACATCCGCTGCAGCGACGGCAAGTTCATCTCGCTCTACGCCATCGTGGACTACTTCAACAACAGGAAGCTGCCCAACTTCACGGACCTCCCGAAGGTCTTCATCTTCCAAGCTTGCAGGGGCGAGAACGAGGACTTCGTGCAGGGCGGCGGAGCGCTGGCGCCGGCGCCGCAGGTGGCGCGGGACGGGGCGGTGCGGGCCCCCCGCGCGCCGCACCACCCCGCGCCCCTGCCGCTGCACCACGCCGAGAAGCCGCTCTACTCCAACATACTCATCGCTAACTCCACCCTCCCAGGGTTCATCTCCCACCGCGACGTGAAGGCCGGCTCCTGGTACATCCAGGTCCTGTGCGAAGTGTTCGCCGAGCGCGCCCACGACTGCCACGTGGACGAGCTGTTCACGCTGGTGGACAAGCGCCTGCGCCTGCGGTTCGGGCGGCAGACCTCCTCCGTGGACCGCTGGGGCTTCAACAACAAGCTGTACTTACACCCCGGTCTGTACGAGTGA
Protein
MTKFYIYLRPRQGSDAPVPPPPVNSSEPVAIPCFHVKKSTHFFEDDVSRDIQLYRTRSRKRGVLVVFSYISFLLNIEPNRDGVNVDCDKLKYLFSEIGFEVLTYQNLTLEQTRATLEQMSSVLAGYECVFLVVSSHGHERRYSSDTDIRCSDGKFISLYAIVDYFNNRKLPNFTDLPKVFIFQACRGENEDFVQGGGALAPAPQVARDGAVRAPRAPHHPAPLPLHHAEKPLYSNILIANSTLPGFISHRDVKAGSWYIQVLCEVFAERAHDCHVDELFTLVDKRLRLRFGRQTSSVDRWGFNNKLYLHPGLYE
Summary
Similarity
Belongs to the peptidase C14A family.
Uniprot
E0D2V3
H9J006
G0XQH2
E9JEG3
A0A2H4WW93
K7WJ03
+ More
A0A2W1BPE7 G0XQH4 A0A2H1WTM5 A0A286M5H6 A0A2A4IVH1 R4I520 A0A0N1I6J1 M4NKN4 A0A194Q5C6 H3JZW6 G0XQH5 A0A212FNV6 A0A023J7Q9 W8DU35 A0A0L7LJ92 A0A1Y1JWU1 K7J0Y7 A0A1S4FTQ4 A0A1W4UN01 A0A232EQK8 A0A182GYM4 Q16PQ5 A0A2J7PU39 K7JH14 A0A1Q3FLD2 A0A1Q3FKV6 A0A2J7PU40 A0A1Q3FL48 A0A1Q3FKY6 B3M583 A0A1Q3FLA8 B4PDY8 A0A0M4EG02 A0A1W4WWI8 A0A182NNC1 B4LGM4 A0A0P4Y7Z4 A0A162DB86 A0A0P6BSY0 A0A0P6IEV2 A0A084WAJ9 B4MLX3 U5EYG7 B4J2U2 A0A182YI42 Q7Q2Q0 A0A182HXI0 A0A182M9Q2 A0A182WZV2 A0A182LI73 A0A182WCK7 A0A2M4AQU3 D6X3B3 A0A182P5Z6 A0A2M4CS65 A0A182FJA8 A0A2M4CSE3 T1GIG7 W5MP74 A0A0P4WPU1 A0A0R3X4P0 V5HTW6 A0A0P4WX74 V9KNT3 B0W757 H3D019 A0A3B3BI00 L7MJV8 A0A3B3BH50 L7MJ43 A0A2D0RE86 A0A3B4FSA4 Q4SC67 A0A212EUQ8 A0A3B3BQT0 A0A3B3D9Y2 A0A2J7PIQ1 A0A0P4VSE4 A0A1V4KP69 A0A1I8NVJ4 T1JVJ5 A0A3B3DJI8 A0A3B3C6K6 A0A2I0M343 A0A2R5LKV7 L7M0M7 A0A1B6E4V5
A0A2W1BPE7 G0XQH4 A0A2H1WTM5 A0A286M5H6 A0A2A4IVH1 R4I520 A0A0N1I6J1 M4NKN4 A0A194Q5C6 H3JZW6 G0XQH5 A0A212FNV6 A0A023J7Q9 W8DU35 A0A0L7LJ92 A0A1Y1JWU1 K7J0Y7 A0A1S4FTQ4 A0A1W4UN01 A0A232EQK8 A0A182GYM4 Q16PQ5 A0A2J7PU39 K7JH14 A0A1Q3FLD2 A0A1Q3FKV6 A0A2J7PU40 A0A1Q3FL48 A0A1Q3FKY6 B3M583 A0A1Q3FLA8 B4PDY8 A0A0M4EG02 A0A1W4WWI8 A0A182NNC1 B4LGM4 A0A0P4Y7Z4 A0A162DB86 A0A0P6BSY0 A0A0P6IEV2 A0A084WAJ9 B4MLX3 U5EYG7 B4J2U2 A0A182YI42 Q7Q2Q0 A0A182HXI0 A0A182M9Q2 A0A182WZV2 A0A182LI73 A0A182WCK7 A0A2M4AQU3 D6X3B3 A0A182P5Z6 A0A2M4CS65 A0A182FJA8 A0A2M4CSE3 T1GIG7 W5MP74 A0A0P4WPU1 A0A0R3X4P0 V5HTW6 A0A0P4WX74 V9KNT3 B0W757 H3D019 A0A3B3BI00 L7MJV8 A0A3B3BH50 L7MJ43 A0A2D0RE86 A0A3B4FSA4 Q4SC67 A0A212EUQ8 A0A3B3BQT0 A0A3B3D9Y2 A0A2J7PIQ1 A0A0P4VSE4 A0A1V4KP69 A0A1I8NVJ4 T1JVJ5 A0A3B3DJI8 A0A3B3C6K6 A0A2I0M343 A0A2R5LKV7 L7M0M7 A0A1B6E4V5
Pubmed
EMBL
AB489117
BAJ16361.1
BABH01018851
HQ328976
AEK20833.1
GQ426280
+ More
ADM32506.1 KY514095 AUD12126.1 JX840448 AFX60235.1 KZ150039 PZC74600.1 HQ328978 AEK20835.1 ODYU01010920 SOQ56332.1 KX827592 ASV64720.1 NWSH01007037 PCG63123.1 JQ768051 AFJ04535.1 KQ460784 KPJ12221.1 JX912275 AGG91491.1 KQ459472 KPJ00210.1 AB663140 BAL60586.1 HQ328979 AEK20836.1 AGBW02004392 OWR55428.1 KF365914 AHH02584.1 KC683711 AHF70712.1 JTDY01000902 KOB75500.1 GEZM01098696 JAV53794.1 NNAY01002753 OXU20628.1 JXUM01021201 KQ560594 KXJ81718.1 CH477774 EAT36368.1 NEVH01021208 PNF19850.1 GFDL01006759 JAV28286.1 GFDL01006911 JAV28134.1 PNF19849.1 GFDL01006832 JAV28213.1 GFDL01006873 JAV28172.1 CH902618 EDV40588.1 GFDL01006751 JAV28294.1 CM000159 EDW93984.1 CP012525 ALC43265.1 CH940647 EDW69462.1 GDIP01232240 JAI91161.1 LRGB01002066 KZS09434.1 GDIP01010428 JAM93287.1 GDIQ01005597 JAN89140.1 ATLV01022201 KE525330 KFB47243.1 CH963847 EDW73184.2 GANO01000312 JAB59559.1 CH916366 EDV96083.1 AAAB01008968 EAA13168.4 APCN01005306 AXCM01001478 GGFK01009819 MBW43140.1 KQ971372 EFA10766.1 GGFL01004006 MBW68184.1 GGFL01004007 MBW68185.1 CAQQ02392379 AHAT01036531 AHAT01036532 AHAT01036533 GDRN01052445 JAI66300.1 UYWX01020493 VDM32914.1 GANP01006785 JAB77683.1 GDRN01052446 JAI66299.1 JW867140 AFO99657.1 DS231852 EDS37662.1 GACK01001560 JAA63474.1 GACK01001731 JAA63303.1 CAAE01014659 CAG01765.1 AGBW02012322 OWR45225.1 NEVH01024980 PNF16221.1 GDKW01001744 JAI54851.1 LSYS01002427 OPJ86218.1 CAEY01000795 AKCR02000044 PKK24100.1 GGLE01006017 MBY10143.1 GACK01007143 JAA57891.1 GEDC01021987 GEDC01009282 GEDC01004338 JAS15311.1 JAS28016.1 JAS32960.1
ADM32506.1 KY514095 AUD12126.1 JX840448 AFX60235.1 KZ150039 PZC74600.1 HQ328978 AEK20835.1 ODYU01010920 SOQ56332.1 KX827592 ASV64720.1 NWSH01007037 PCG63123.1 JQ768051 AFJ04535.1 KQ460784 KPJ12221.1 JX912275 AGG91491.1 KQ459472 KPJ00210.1 AB663140 BAL60586.1 HQ328979 AEK20836.1 AGBW02004392 OWR55428.1 KF365914 AHH02584.1 KC683711 AHF70712.1 JTDY01000902 KOB75500.1 GEZM01098696 JAV53794.1 NNAY01002753 OXU20628.1 JXUM01021201 KQ560594 KXJ81718.1 CH477774 EAT36368.1 NEVH01021208 PNF19850.1 GFDL01006759 JAV28286.1 GFDL01006911 JAV28134.1 PNF19849.1 GFDL01006832 JAV28213.1 GFDL01006873 JAV28172.1 CH902618 EDV40588.1 GFDL01006751 JAV28294.1 CM000159 EDW93984.1 CP012525 ALC43265.1 CH940647 EDW69462.1 GDIP01232240 JAI91161.1 LRGB01002066 KZS09434.1 GDIP01010428 JAM93287.1 GDIQ01005597 JAN89140.1 ATLV01022201 KE525330 KFB47243.1 CH963847 EDW73184.2 GANO01000312 JAB59559.1 CH916366 EDV96083.1 AAAB01008968 EAA13168.4 APCN01005306 AXCM01001478 GGFK01009819 MBW43140.1 KQ971372 EFA10766.1 GGFL01004006 MBW68184.1 GGFL01004007 MBW68185.1 CAQQ02392379 AHAT01036531 AHAT01036532 AHAT01036533 GDRN01052445 JAI66300.1 UYWX01020493 VDM32914.1 GANP01006785 JAB77683.1 GDRN01052446 JAI66299.1 JW867140 AFO99657.1 DS231852 EDS37662.1 GACK01001560 JAA63474.1 GACK01001731 JAA63303.1 CAAE01014659 CAG01765.1 AGBW02012322 OWR45225.1 NEVH01024980 PNF16221.1 GDKW01001744 JAI54851.1 LSYS01002427 OPJ86218.1 CAEY01000795 AKCR02000044 PKK24100.1 GGLE01006017 MBY10143.1 GACK01007143 JAA57891.1 GEDC01021987 GEDC01009282 GEDC01004338 JAS15311.1 JAS28016.1 JAS32960.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000002358 UP000192221 UP000215335 UP000069940 UP000249989 UP000008820 UP000235965 UP000007801 UP000002282 UP000092553 UP000192223 UP000075884 UP000008792 UP000076858 UP000030765 UP000007798 UP000001070 UP000076408 UP000007062 UP000075840 UP000075883 UP000076407 UP000075882 UP000075920 UP000007266 UP000075885 UP000069272 UP000015102 UP000018468 UP000046396 UP000274429 UP000002320 UP000007303 UP000261560 UP000221080 UP000261460 UP000190648 UP000095300 UP000015104 UP000053872
UP000002358 UP000192221 UP000215335 UP000069940 UP000249989 UP000008820 UP000235965 UP000007801 UP000002282 UP000092553 UP000192223 UP000075884 UP000008792 UP000076858 UP000030765 UP000007798 UP000001070 UP000076408 UP000007062 UP000075840 UP000075883 UP000076407 UP000075882 UP000075920 UP000007266 UP000075885 UP000069272 UP000015102 UP000018468 UP000046396 UP000274429 UP000002320 UP000007303 UP000261560 UP000221080 UP000261460 UP000190648 UP000095300 UP000015104 UP000053872
Pfam
PF00619 CARD
Interpro
ProteinModelPortal
E0D2V3
H9J006
G0XQH2
E9JEG3
A0A2H4WW93
K7WJ03
+ More
A0A2W1BPE7 G0XQH4 A0A2H1WTM5 A0A286M5H6 A0A2A4IVH1 R4I520 A0A0N1I6J1 M4NKN4 A0A194Q5C6 H3JZW6 G0XQH5 A0A212FNV6 A0A023J7Q9 W8DU35 A0A0L7LJ92 A0A1Y1JWU1 K7J0Y7 A0A1S4FTQ4 A0A1W4UN01 A0A232EQK8 A0A182GYM4 Q16PQ5 A0A2J7PU39 K7JH14 A0A1Q3FLD2 A0A1Q3FKV6 A0A2J7PU40 A0A1Q3FL48 A0A1Q3FKY6 B3M583 A0A1Q3FLA8 B4PDY8 A0A0M4EG02 A0A1W4WWI8 A0A182NNC1 B4LGM4 A0A0P4Y7Z4 A0A162DB86 A0A0P6BSY0 A0A0P6IEV2 A0A084WAJ9 B4MLX3 U5EYG7 B4J2U2 A0A182YI42 Q7Q2Q0 A0A182HXI0 A0A182M9Q2 A0A182WZV2 A0A182LI73 A0A182WCK7 A0A2M4AQU3 D6X3B3 A0A182P5Z6 A0A2M4CS65 A0A182FJA8 A0A2M4CSE3 T1GIG7 W5MP74 A0A0P4WPU1 A0A0R3X4P0 V5HTW6 A0A0P4WX74 V9KNT3 B0W757 H3D019 A0A3B3BI00 L7MJV8 A0A3B3BH50 L7MJ43 A0A2D0RE86 A0A3B4FSA4 Q4SC67 A0A212EUQ8 A0A3B3BQT0 A0A3B3D9Y2 A0A2J7PIQ1 A0A0P4VSE4 A0A1V4KP69 A0A1I8NVJ4 T1JVJ5 A0A3B3DJI8 A0A3B3C6K6 A0A2I0M343 A0A2R5LKV7 L7M0M7 A0A1B6E4V5
A0A2W1BPE7 G0XQH4 A0A2H1WTM5 A0A286M5H6 A0A2A4IVH1 R4I520 A0A0N1I6J1 M4NKN4 A0A194Q5C6 H3JZW6 G0XQH5 A0A212FNV6 A0A023J7Q9 W8DU35 A0A0L7LJ92 A0A1Y1JWU1 K7J0Y7 A0A1S4FTQ4 A0A1W4UN01 A0A232EQK8 A0A182GYM4 Q16PQ5 A0A2J7PU39 K7JH14 A0A1Q3FLD2 A0A1Q3FKV6 A0A2J7PU40 A0A1Q3FL48 A0A1Q3FKY6 B3M583 A0A1Q3FLA8 B4PDY8 A0A0M4EG02 A0A1W4WWI8 A0A182NNC1 B4LGM4 A0A0P4Y7Z4 A0A162DB86 A0A0P6BSY0 A0A0P6IEV2 A0A084WAJ9 B4MLX3 U5EYG7 B4J2U2 A0A182YI42 Q7Q2Q0 A0A182HXI0 A0A182M9Q2 A0A182WZV2 A0A182LI73 A0A182WCK7 A0A2M4AQU3 D6X3B3 A0A182P5Z6 A0A2M4CS65 A0A182FJA8 A0A2M4CSE3 T1GIG7 W5MP74 A0A0P4WPU1 A0A0R3X4P0 V5HTW6 A0A0P4WX74 V9KNT3 B0W757 H3D019 A0A3B3BI00 L7MJV8 A0A3B3BH50 L7MJ43 A0A2D0RE86 A0A3B4FSA4 Q4SC67 A0A212EUQ8 A0A3B3BQT0 A0A3B3D9Y2 A0A2J7PIQ1 A0A0P4VSE4 A0A1V4KP69 A0A1I8NVJ4 T1JVJ5 A0A3B3DJI8 A0A3B3C6K6 A0A2I0M343 A0A2R5LKV7 L7M0M7 A0A1B6E4V5
PDB
3J9K
E-value=1.8868e-26,
Score=295
Ontologies
PATHWAY
GO
GO:0006915
GO:0004197
GO:0042981
GO:0097200
GO:0022416
GO:0043293
GO:0010940
GO:0007291
GO:0097199
GO:2001269
GO:0010506
GO:0048749
GO:0006919
GO:0048813
GO:0046672
GO:0035234
GO:0008258
GO:0016540
GO:0007516
GO:0007417
GO:0016322
GO:0035006
GO:0051291
GO:0008340
GO:0031638
GO:0042803
GO:0035070
GO:0045476
GO:0008285
GO:0005737
GO:0097153
GO:0043065
GO:0008630
GO:0043281
GO:0097205
GO:0048696
GO:0001822
GO:0006508
GO:0008234
GO:0008026
GO:0016818
GO:0004003
GO:0003677
GO:0000723
GO:0005634
GO:0006260
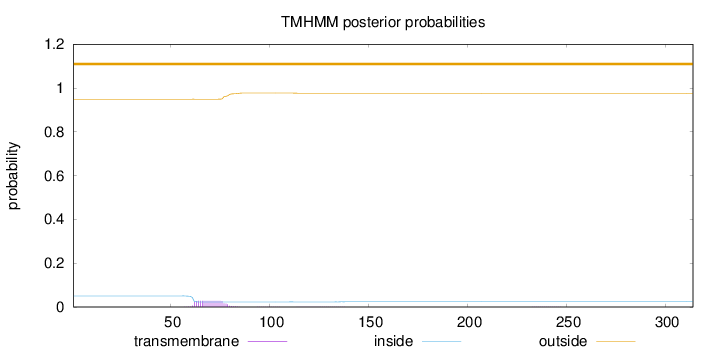
Topology
Length:
314
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.51504
Exp number, first 60 AAs:
0.00426
Total prob of N-in:
0.05072
outside
1 - 314
Population Genetic Test Statistics
Pi
5.016642
Theta
14.113231
Tajima's D
-1.510562
CLR
17.352465
CSRT
0.056347182640868
Interpretation
Uncertain
