Gene
KWMTBOMO06045 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002840
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_translational_activator_GCN1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.544 Nuclear Reliability : 1.231
Sequence
CDS
ATGGCTGATACAGAGGTATTAAAGAAATTAAAAGATATTCCATTTAAAATCCAGACAGCAAGTTTAAAAGGGAGACTTGAAGTAGTTAATGAAATACGTGAAATTTTATCGTCGCCAGGTATAACAGAACCAGCAGTAAAGTTTGTTTGCCGAGTGCTAGCTCTGACATTGCACAGATATAGAGATACAACGTCCCAATCTTACATCAAGAGTCTTGTGGCTTTCCTTGCTACTGAGCACACAGAATGGGCATTTAAGAATTTATCACCCGTATTACTAGAGATATCAGAACAGCTGAAAAACAACTCAACCTCAAAAAGCAACAGCCAGTCAGGTCTGTTTGCACTTCGATGGTCTAGTGTACTGGTTGAGTCTTCACCGAAATCCTTAGAGCATGAAGTGGACCTGAACTCATTGATACTTGCGCAGGCTAATTTACTAGCCACTGTTTGTGCATATGGCGATAAGAAGAAGAATAAAAAAGCTTATTCTGTGCTACATGCGGTATGGAGTAAAATAGGAAAAGAGAAAGTACTGAAATGGGTGGAAACATTACTAGCACAGCCTGGTGACAGCGGACCGCACATCTCAGTGACATTCTCTGCTCTCTGTGCTCATTATAAGGATATAGGAGATGAATCTATTATAACATTACATAAGGCCAAAATGTTAGAAAATTTTACTAAAAGTCTGATCAGCGTGAAGTCCAGGCCAAATGCGAACTACATCAAGGGCTGCTCTGACCTCCTGCACCTGCTCAGCGAGTCTGATATCCAAGACACATTACTGCCGGCTCTGCACAAAGCCATGCTCCGGAGTCCGGAAACGATCGTTCAAGCTGTCGGAGAAGTGGTTGCCAATCTCAATATCTACCTGGATGGCGTGGCTGTTGATATCGGAAAGAGTTTGATAGTGAATCTCCATTCTAAAGACGAGTGGGTCCGCGCGGAGGCGCTGAGTGCGCTGCACCGGCTGGCGGCGCGCACCCGCACCGCGCCCGCCGCCGCCGCGCTGCTCGCGCACGCATTCGCGGAGTACAACGGCGCCAGCGGGAAACTCACTTCCAGCGAAGATAAGATTGCAGTTCTCAATGGAGTACGCGCCCTGAGCCAGCTGGCCGTCGCCCCGGAGGACAGACGGGCGCTGTTCGCGGAGACGGCCGCCCACACCGCGCGCATACTGGACTCCGAGTCCCACGAGCGAACCTTGTGCGTGGCCCTGGACGTACTGCAGCACTGGACCACAGGCCTGGATGGACCGCTGCCCGATGTCATTCTTGAGATATTCACGAAGAACATATCGGCGAAGAGCAGCACGCAGACCGTCCGCACGGCGTACACCAGCCTGCTGGGCCGCGCGCTGCGCCGCTGCGGGCCGCCCGCCGCCGCCGCGCCGCTGCACGCGCTGCTGCTGCAGCCCGTCGAGAGGGCCGCGCTGCAGCCGCTGCAGAACGCCACAGTAAGCGAGGGTATCTGCGCGATGCTGGGCCTGGTGCTGCTGGACGAGTCCCGGCAGCTGCCCGGCAAGTCCTCGTGCTGGGCCGCCCTGCTGCACCCGGACAAGCACGTGCTGCTGCACGACAGGGTGCTCGCCGCCGCATCCGAACACGTCCTCATACAGGTGGCGTTACTGTGCAGAACGGTGTTGGACCACTACCCGAGTGTTGCCAGCGAACCAAACTCCCCGATCTACAGAGCCTTCATTATGGTCCTACTGTCTACCGTGAAGCCAGTTCGAGAAGTAGCACTGCAAGAAGTCAGAGCTTTACTCGCAAAGCGAGATAAAGCGGCAACGGCCAAGCACTTAGTGCTGAAGCTGAACGAAGTTCTGGATGAAGGAAAAATCTTCCATATTAAAGAAAAGTCCCCCTCGGACGAGAAAGGAGCTGAGGTCACCGGGAAGATGATACTAGACTGCGCGCATGCTCTTTGCTCTTATAAAGACTATCCCAAAGAAGAGCTGGAGAGCATCGCGCTGTCGGCGTTGCCGTGCAGCCACCACCCGTTGGTAAAGGCTGGAGCTCCACGAGCCTGGGTGAAGATGGTCTCATCACTCGGTCTGCAGCCGAAAGAGCTGGTCACTCTTTATGCCAATAACTTGAAGAATACATACATCAACGGCTTTACGCCTAGTGAGACTGTGTCGAGCGTAGTGGAAACGCTGGTGTCGGTGGGCCCCGACGGCATGTCGTCGAGCGCCGTGCAGGCGGCGCTGGTGTGGCTCCAGGACCCGCAGCTGCAGCGCGTCACCAGGGACGAGTACTTCGTCTACCTCACGCCAGACGACGACATCTATGACAAGTCCAGCGTGCCCGGTAATGAAGACAACAAAGAGATGAATTTGAAACGAGAGAGCAAAGCGTACAGCTACAAGGAGCAGCTCGAAGAGTTGCAGCTGCGCCGAGAGCTGGACGAGAAGAAACGTAAAGAGGGGAAAATTAAGGAGACGCCGTTATCTGCTAAACAAAAAGAAGCCATAAAAGCGCAACTGGCCAAGGAAAGTGCGGTCAGGAAGAGGCTCAGTGTGTTGAACGAACGTTTAGTTTGCGCCGTGGGTCTGCTGGAGGCGGTCCAGCGAGGCTGCCCCTCCGCCCTGCGCAGCAGTGCGCGGCGCGTGGTGTCCCGCGTGCTGGCGGCGCTGCGCAGTCCGCTGGCGGCGCCGCGCCTGCTGCCCGCGCACCGCCGCCTGGGCGCCGCGCTGCTGCCCGCGCGCCTGCCCGCCCCGCTGCGCGACCACGTCACCCGCACCACGCTCAGGCTCTATAAACCTCAATGCGACCTAGACCCGAGCTGGGAAGACGAGGACCTGGTTGAAGCCACCTCGCGAGTCATCAACCTGCTGCACGCCGCCAGCGTGGCCGTGAAGGTGGATGCCAGTGACACCAAAAAGTTAAACCATTTTACCGCTCCCGGATTCTGCTACGTCTTTCCTGTACTCAAGTTAGGTTTAACTAAAGAACTGAGTGAAAAGGACGAAATGACAATGTTGAACGGGCTGGCGGTGATCACGCACCACGTGTCGGCGCGGCCCCACCCGGACGACGCCGAGCTGCTGCACCCGGCGCTGTTCCCCATGGACGAGATATTCCGACTGCTCATAGAACTCATCGAGAGCACGCGGCTGTGCGCGCAGGCGGCCTGCATCGTGGCGCTGCTGGAGGCGGCCCGGTGCGCGGCGCGGGGGCCCCTCGCGCGGGACGACGCCGCCGCGCTGCTGGCCGGCCTGCGCAGCGAGAGCGAGGTGGTCCGCGACACGGTGCTGCAGGCGCTGCTGAGCCTGGCGGAGCGGCTGCCGCCCCTGCTGCAAGACGAAGACTACGCCTACTCCGTCGTCAAGCGGATCCTGATCGCCACCTTCGACGTAGTTGAAGACAACAAGAAGCTAGCCGTGGAGCTGTGGGAGCGCGTGCCCGGCTGCGAGCCCGTGTCCCGCGGCGCGCTGATGGCGGACCTGGTGCTGGACGAGCTGCAGACCGCCTGTCTCCCGCTCCAGCGAGCAGCAGCCCTCGCCCTGGCGCACCTCGTCCTCCGCGGAGACGCTCCGTGCGACGTCCTGCTGCAGCGTCTACAGGACCTGTACGCTGAAAAGCTGCCCCTGATCCCGGCCAAGCAAGACCAGTTCGGTCACGAGGTGGAGCCGGCGCTGGACATGTGGGAGGGGCGGCGCGGCGTGGCGCTGGGGCTGGGCGCGCTGGCCCCGCGGCTGCAGGTGGGCGCGGTGCCGGCCGTCATGCGCTTCTTCGTGGAGTGGGGCCTGGCCGACCGCCGCGACGACGTGCGCGCCGAGATGCTCGCCGCCGCCATGGCCATCGTCGAGCTGCACGGGAAGGATACGATCAGCAGCCAGCTGCCGGTGTTCGAGAGGTTCCTGGACTCCGCGCCCAAGTCTGGCAGCTACGACGCGGTGCGGGAGGCCGTGGTGCTGCTGGTGGGGTCCCTGGCGCGCCACCTGGAGCCCGACGACGTGCGCCTGCGCCCCATCGCGCTGCGCCTCGTCGCCGCCCTGTCCACGCCGGCTCAGCAGGTCCAGGAGGCGGTGGGCAACTGTCTGCCGCACCTGGTGACGTCACCGGCGCTGGAGAGCGAGATCCCGGGCATCGTCAACAAGCTGATGAAACAACTACTCACCGCCGAGAAATTCGGAGATCGAAGGGGCGCCGCGTACGGCATAGCCGGCGTCGTCAAGGGGCTCGGCATCCTGTCCCTGAAGCAGCTGGACATCATGGGGAAACTCACAGAAGCCATACAGGAAAAGAAGAACTACAAGTATCGCGAAGGCGCGCTGTTCGGGTTCGAGCTGCTGTGCCGCGTGCTGGGCCGCCTGTTCGAGCCGTACATCGTGCACGTGCTGCCGCACCTGCTGCTGTGTCTGGGCGACGGCTCGCAGTACGTGCGCACCGCCGCCGACGACACCGCGCGCCTCATCATGAGCCGCCTCAGCGCGCACGGCGTCAAGCTGGTGCTGCCCGCCCTGCTCAACGCCGTCAACGACGACAACTGGAGGACTAAAGCGGGGTCCATCGAGCTGCTGGGCGCCATGGCGTACTGCGCGCCCAAGCAGCTGTCGTCGTGCCTGCCGTCCATCGTGCCGCGCCTCATCGAGGTGCTGAGCGACTCGCATCTGCGCGTGCAGCTGTCCGCCGCCGCCGCGCTCAAGGTCATCGGCTCCGTCATCAGGAACCCCGAGATCCAAGCCATCGTGCCGGTGCTGCTGCAAGCCCTGCAGGACCCGTCCAACAAGACGTCGGCGTGTCTGCAGACGCTGCTGGACACCAAGTTCGTCCACTTCATCGACGCCCCGTCGCTGGCCCTCATCATGCCGGTGGTGCAGCGAGCCTTCCTGGACCGGAGCACCGAGACCCGCAAGATGGCGGCGCAGATCATAGGCAACATGTACTCGCTGACCGACCAGAAGGACCTCACACCCTATCTCCCGAACATCATCCCGGGACTGAAAAGCTCTCTACTGGATCCCGTGCCGGAGGTCCGCAGCGTCTCGGCGCGAGCCCTCGGCGCTATGGTAAAGGGTATGGGAGAGAGCAGCTTCGAAGAGTTGCTCCCGTGGCTGATGCACACGCTCACCTCGGAGACCAGCTCCGTGGACCGCTCCGGTGCTGCCCAGGGACTCTCCGAAGTCGTCGCTGGACTTGGAGTGCAAAAATTGCACAAGATTATGCCAGACATCATCGCCACGGCGGAGCGCACGGACATCGCGCCGCACGTCAAGGACGGCTACATCATGATGTTCATCTACATGCCCGGCGCCTTCACCGACGAGTTCACCCCCTACATCGGCCAGATCATCAACCCCATCCTCAAGGCGCTGGCGGACGAGAACGAGTACGTGCGCGAGACGGCGCTGCGGGCCGGCCAGCGCATCGTCAACCTGTACGCGGAGTCCGCCATCGCGCTGCTGCTGCCAGAGCTCGAGAAGGGCCTGTTCGACGACAACTGGAGGATCCGCTACAGCTCCGTGCAGCTGCTGGGCGACCTGCTGTACCGCATCTCCGGGGTCACCGGCAAGATGAGCACCGAGACCGCCTCGGAGGACGACAACTTCGGCACGGAGCAGTCGCACAAGGCCATCATCACCGCGCTGGGCCCCGAGCGCAGGAACCGCGTGCTGGCCGGCCTCTACATGGGCCGCAGCGACGTCGCGCTCATGGTGCGCCAGGCCGCGCTGCACGTCTGGAAGGTGGTCGTCACCAACACTCCCAAGACGTTACGCGAGATCCTGCCGACCCTCTTCGGGTTGCTGCTCGGATGCCTCGCCTCCACCAGCTACGACAAGCGACAGGTTGCCGCCAGGACCCTGGGAGATCTCGTCAGGAAGCTGGGCGAGCGCGTGCTGCCGGAGATAGTGCCCATCCTGGAGCGCGGGCTGCGGTCTCCGCGCGCCGACCAGCGCCAGGGCGTCTGCATCGGGCTCGGGGAGATCCTCGCCTCCACCTCGCGGGACGCCGTGCTCAGCTTCGCCGACGGATTGGTGCCCACGGTCCGCACCGCGCTGTGCGACCCGCTGCCGGAGGTGCGGCTGGCTGCGGCGCGCACGTTCGACAGTCTGCACGCCACCATCGGCAACAAGGCGCTGGACGACATCCTGCCCTACATGCTCGAGGGCCTGAAGGACCCCGACCCCGCCGTCGCGGACGCCACGCTCGACGGACTCAAACAGATAATGTCGATAAAGTCGCGCGCGGTGCTGCCCTACCTGATCCCGGTGCTGACGGGCGGCGGCGGCGGCGGCGTGGACACGCGCGCGCTGTCGGCGCTGGCGGCGGCGGCCGGCGGGGCCCTGGCGCGCCACGTGCCGCGCGTGCTGCCCGCGCTGCTGCACGCGCTGGAGGCCGCGCGCGCCACGCCGCACGAGGCCGCCGAGCTGGACCACTGCCGCGACGCGCTGCTGCCCGTCACCGACCCGCCGGGACTGCGATGCATTATCGACACGCTGCTGGACACGGTCCGCTCCAGCGAGGGCTCCCGGCGCCGCGCCGCCGCCGTGCTGCTGTGCGCCTTCGTCGCGCACACCCGCGGCGACCTAGAGCCGCACGTGCCGCAGCTGCTGCGGGGCCTGGTGCTGCTGCTGGCCGACGACGACCGAGACGTGCTGCTCATGGCGTGGGAGGCGCTCTCCGCGCTCACCAAGACGCTGGACGCCGAGCGGCAGATCCAGCACGTGTCCGACGTGCGCCAGGCCGTGCGCTATGCGGCCGCCGACCTCAAGGGGGATCTCTTGCCCGGATTCTGTCTGCCCAAGGGTATAGCGCCGATCCTGCCTCTGTTCCGGGAGTCTATCCTAAACGGCTTACCTGAAGACAAGGAAAATGCGGCGCTGATGCTGGGCGAGGTCATCAAGCTGACGTCTCCGGCCGCCATACAGCCCTCCGTCGTACACATCACCGGACCTCTCATCCGTATCCTCGGAGACAGGTTCAACTCTAGCGTCAAAGCTGCCGTTTTGGAGACCTTAGCCTCTTTACTCTCGAAGGTCGGGGTAATGCTGAAGCAGTTCTTACCGCAACTGCAGACGACGTTCCTGAAGGCCCTCAACGATCCCAACCGGCCCGTGCGGATCAAGGCGGGCCTGGCTCTGTCACAACTGGTCATCATCCACACCAGAGCGGATCCGCTCTTTGTGGAGATGCACAACGGCATTAAGAATACAGACGACCCCGCCATAAGGGAGACCATGCTGCAAGCGTTGCGCAGCATCATCTCCGGCGGCGGAGACAAGCTATCGGAGCAACTGGCGCTCAGTATTCTGTCGACGCTGACGAGCCCCGCGCTCCTGGCGCACCCGGAGGACCCGCCCCGGGGAGCGGTGGGGGGCTGTCTGGGCGCCCTGCTGCACTGGTTGCCGTCCGCACACCACGACGCTGCCCTGCAACACCACGTCTTGGCACAATCCGACGACTGGCTGCTCCAACACGGCCGCTCCTGTGCTCTCTTCGTGGCACTCAAGGAGACCCCGGCCTCTATATACCGCGATCATTACGAAGAAAAGATAGACAAAACCCTGTTGGCGTACCTGGCCAGCGACAAGATCCCGATCGTCTGTAACGGTATCCGCGGCATCGGCTTCCTCATGAAGTATCTCCTCCAGAACGACAGGCCGCTCCCACAGAACATACTTTCGCAATTTGTCAGAAGCATGAACCATCAAAGCAACGAGGTGAAGCAGATGATGGCGCGGGCCAGCGGCGCCCTGGGTCGCGAGGCCGACAACCCGGAGCTCCTGCGTGCCGTGCTGCCCGCCCTCGTCAACGGCACCAAGGAGAAGAACTCGTACGTGCGCGCCAACGCGGAGTCCTCGCTCCGAACCCTGCTGCGGCTTCATTCTGAACCAGACTATTTCCAGCGATGCGTGTCGTTCCTGGAGGAAGGCGCCCGTGAAGCTCTGTCTGATGTAGTAGCGAGAGTATTGCGTCGACCTCAACCCGACGGACGAGATGAGGACCTGGACTGTACGCTCATCACCTGA
Protein
MADTEVLKKLKDIPFKIQTASLKGRLEVVNEIREILSSPGITEPAVKFVCRVLALTLHRYRDTTSQSYIKSLVAFLATEHTEWAFKNLSPVLLEISEQLKNNSTSKSNSQSGLFALRWSSVLVESSPKSLEHEVDLNSLILAQANLLATVCAYGDKKKNKKAYSVLHAVWSKIGKEKVLKWVETLLAQPGDSGPHISVTFSALCAHYKDIGDESIITLHKAKMLENFTKSLISVKSRPNANYIKGCSDLLHLLSESDIQDTLLPALHKAMLRSPETIVQAVGEVVANLNIYLDGVAVDIGKSLIVNLHSKDEWVRAEALSALHRLAARTRTAPAAAALLAHAFAEYNGASGKLTSSEDKIAVLNGVRALSQLAVAPEDRRALFAETAAHTARILDSESHERTLCVALDVLQHWTTGLDGPLPDVILEIFTKNISAKSSTQTVRTAYTSLLGRALRRCGPPAAAAPLHALLLQPVERAALQPLQNATVSEGICAMLGLVLLDESRQLPGKSSCWAALLHPDKHVLLHDRVLAAASEHVLIQVALLCRTVLDHYPSVASEPNSPIYRAFIMVLLSTVKPVREVALQEVRALLAKRDKAATAKHLVLKLNEVLDEGKIFHIKEKSPSDEKGAEVTGKMILDCAHALCSYKDYPKEELESIALSALPCSHHPLVKAGAPRAWVKMVSSLGLQPKELVTLYANNLKNTYINGFTPSETVSSVVETLVSVGPDGMSSSAVQAALVWLQDPQLQRVTRDEYFVYLTPDDDIYDKSSVPGNEDNKEMNLKRESKAYSYKEQLEELQLRRELDEKKRKEGKIKETPLSAKQKEAIKAQLAKESAVRKRLSVLNERLVCAVGLLEAVQRGCPSALRSSARRVVSRVLAALRSPLAAPRLLPAHRRLGAALLPARLPAPLRDHVTRTTLRLYKPQCDLDPSWEDEDLVEATSRVINLLHAASVAVKVDASDTKKLNHFTAPGFCYVFPVLKLGLTKELSEKDEMTMLNGLAVITHHVSARPHPDDAELLHPALFPMDEIFRLLIELIESTRLCAQAACIVALLEAARCAARGPLARDDAAALLAGLRSESEVVRDTVLQALLSLAERLPPLLQDEDYAYSVVKRILIATFDVVEDNKKLAVELWERVPGCEPVSRGALMADLVLDELQTACLPLQRAAALALAHLVLRGDAPCDVLLQRLQDLYAEKLPLIPAKQDQFGHEVEPALDMWEGRRGVALGLGALAPRLQVGAVPAVMRFFVEWGLADRRDDVRAEMLAAAMAIVELHGKDTISSQLPVFERFLDSAPKSGSYDAVREAVVLLVGSLARHLEPDDVRLRPIALRLVAALSTPAQQVQEAVGNCLPHLVTSPALESEIPGIVNKLMKQLLTAEKFGDRRGAAYGIAGVVKGLGILSLKQLDIMGKLTEAIQEKKNYKYREGALFGFELLCRVLGRLFEPYIVHVLPHLLLCLGDGSQYVRTAADDTARLIMSRLSAHGVKLVLPALLNAVNDDNWRTKAGSIELLGAMAYCAPKQLSSCLPSIVPRLIEVLSDSHLRVQLSAAAALKVIGSVIRNPEIQAIVPVLLQALQDPSNKTSACLQTLLDTKFVHFIDAPSLALIMPVVQRAFLDRSTETRKMAAQIIGNMYSLTDQKDLTPYLPNIIPGLKSSLLDPVPEVRSVSARALGAMVKGMGESSFEELLPWLMHTLTSETSSVDRSGAAQGLSEVVAGLGVQKLHKIMPDIIATAERTDIAPHVKDGYIMMFIYMPGAFTDEFTPYIGQIINPILKALADENEYVRETALRAGQRIVNLYAESAIALLLPELEKGLFDDNWRIRYSSVQLLGDLLYRISGVTGKMSTETASEDDNFGTEQSHKAIITALGPERRNRVLAGLYMGRSDVALMVRQAALHVWKVVVTNTPKTLREILPTLFGLLLGCLASTSYDKRQVAARTLGDLVRKLGERVLPEIVPILERGLRSPRADQRQGVCIGLGEILASTSRDAVLSFADGLVPTVRTALCDPLPEVRLAAARTFDSLHATIGNKALDDILPYMLEGLKDPDPAVADATLDGLKQIMSIKSRAVLPYLIPVLTGGGGGGVDTRALSALAAAAGGALARHVPRVLPALLHALEAARATPHEAAELDHCRDALLPVTDPPGLRCIIDTLLDTVRSSEGSRRRAAAVLLCAFVAHTRGDLEPHVPQLLRGLVLLLADDDRDVLLMAWEALSALTKTLDAERQIQHVSDVRQAVRYAAADLKGDLLPGFCLPKGIAPILPLFRESILNGLPEDKENAALMLGEVIKLTSPAAIQPSVVHITGPLIRILGDRFNSSVKAAVLETLASLLSKVGVMLKQFLPQLQTTFLKALNDPNRPVRIKAGLALSQLVIIHTRADPLFVEMHNGIKNTDDPAIRETMLQALRSIISGGGDKLSEQLALSILSTLTSPALLAHPEDPPRGAVGGCLGALLHWLPSAHHDAALQHHVLAQSDDWLLQHGRSCALFVALKETPASIYRDHYEEKIDKTLLAYLASDKIPIVCNGIRGIGFLMKYLLQNDRPLPQNILSQFVRSMNHQSNEVKQMMARASGALGREADNPELLRAVLPALVNGTKEKNSYVRANAESSLRTLLRLHSEPDYFQRCVSFLEEGAREALSDVVARVLRRPQPDGRDEDLDCTLIT
Summary
Uniprot
H9J005
A0A0N1INW4
A0A194Q3U1
A0A212FNW4
A0A195DI45
A0A158NQR2
+ More
A0A195BE87 A0A151WN86 E9IG54 A0A195D3P1 A0A067RTH7 A0A2J7PU34 A0A026WLG5 Q177C0 A0A1S4FCV3 E0V8V7 A0A182H7A1 W5JJ28 A0A182IZ36 A0A182RIM2 A0A2M4B849 A0A182F7I1 A0A182MBV7 A0A182Y3R4 A0A084VD37 A0A182NRV3 A0A182VZK6 A0A182QX04 A0A182PSK4 A0A182TWJ7 Q7PY15 A0A2M4A3T0 A0A182WSR7 A0A182HLM0 A0A182JRZ4 A0A182VKQ5 A0A195FSL9 T1HJQ7 A0A0L0CLP2 A0A1L8DRD4 A0A2M4CMG6 W8CCK9 A0A2M4CPJ6 A0A1I8NZR9 T1J2L3 A0A3B0JLZ1 A0A023GMF5 A0A224Z218 L7LXL1 A0A2R5LKZ7 I3KG96 I3KG97 A0A3Q3N3M1
A0A195BE87 A0A151WN86 E9IG54 A0A195D3P1 A0A067RTH7 A0A2J7PU34 A0A026WLG5 Q177C0 A0A1S4FCV3 E0V8V7 A0A182H7A1 W5JJ28 A0A182IZ36 A0A182RIM2 A0A2M4B849 A0A182F7I1 A0A182MBV7 A0A182Y3R4 A0A084VD37 A0A182NRV3 A0A182VZK6 A0A182QX04 A0A182PSK4 A0A182TWJ7 Q7PY15 A0A2M4A3T0 A0A182WSR7 A0A182HLM0 A0A182JRZ4 A0A182VKQ5 A0A195FSL9 T1HJQ7 A0A0L0CLP2 A0A1L8DRD4 A0A2M4CMG6 W8CCK9 A0A2M4CPJ6 A0A1I8NZR9 T1J2L3 A0A3B0JLZ1 A0A023GMF5 A0A224Z218 L7LXL1 A0A2R5LKZ7 I3KG96 I3KG97 A0A3Q3N3M1
Pubmed
EMBL
BABH01018852
BABH01018853
BABH01018854
KQ460784
KPJ12223.1
KQ459472
+ More
KPJ00208.1 AGBW02004392 OWR55425.1 KQ980824 KYN12565.1 ADTU01023474 KQ976511 KYM82524.1 KQ982911 KYQ49326.1 GL762910 EFZ20480.1 KQ976885 KYN07471.1 KK852473 KDR23119.1 NEVH01021208 PNF19843.1 KK107154 EZA56885.1 CH477378 EAT42272.1 DS234986 EEB09813.1 JXUM01115842 JXUM01115843 JXUM01115844 JXUM01115845 KQ565921 KXJ70533.1 ADMH02001371 ETN62794.1 GGFJ01000029 MBW49170.1 AXCM01000082 ATLV01011143 KE524645 KFB35881.1 AXCN02000250 AAAB01008987 EAA00880.4 GGFK01001977 MBW35298.1 APCN01000909 KQ981280 KYN43458.1 ACPB03016346 JRES01000230 KNC33176.1 GFDF01005147 JAV08937.1 GGFL01002354 MBW66532.1 GAMC01003516 JAC03040.1 GGFL01002650 MBW66828.1 JH431806 OUUW01000007 SPP83265.1 GBBM01000404 JAC35014.1 GFPF01012752 MAA23898.1 GACK01009460 JAA55574.1 GGLE01006078 MBY10204.1 AERX01019889 AERX01019890
KPJ00208.1 AGBW02004392 OWR55425.1 KQ980824 KYN12565.1 ADTU01023474 KQ976511 KYM82524.1 KQ982911 KYQ49326.1 GL762910 EFZ20480.1 KQ976885 KYN07471.1 KK852473 KDR23119.1 NEVH01021208 PNF19843.1 KK107154 EZA56885.1 CH477378 EAT42272.1 DS234986 EEB09813.1 JXUM01115842 JXUM01115843 JXUM01115844 JXUM01115845 KQ565921 KXJ70533.1 ADMH02001371 ETN62794.1 GGFJ01000029 MBW49170.1 AXCM01000082 ATLV01011143 KE524645 KFB35881.1 AXCN02000250 AAAB01008987 EAA00880.4 GGFK01001977 MBW35298.1 APCN01000909 KQ981280 KYN43458.1 ACPB03016346 JRES01000230 KNC33176.1 GFDF01005147 JAV08937.1 GGFL01002354 MBW66532.1 GAMC01003516 JAC03040.1 GGFL01002650 MBW66828.1 JH431806 OUUW01000007 SPP83265.1 GBBM01000404 JAC35014.1 GFPF01012752 MAA23898.1 GACK01009460 JAA55574.1 GGLE01006078 MBY10204.1 AERX01019889 AERX01019890
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000078492
UP000005205
+ More
UP000078540 UP000075809 UP000078542 UP000027135 UP000235965 UP000053097 UP000008820 UP000009046 UP000069940 UP000249989 UP000000673 UP000075880 UP000075900 UP000069272 UP000075883 UP000076408 UP000030765 UP000075884 UP000075920 UP000075886 UP000075885 UP000075902 UP000007062 UP000076407 UP000075840 UP000075881 UP000075903 UP000078541 UP000015103 UP000037069 UP000095300 UP000268350 UP000005207 UP000261640
UP000078540 UP000075809 UP000078542 UP000027135 UP000235965 UP000053097 UP000008820 UP000009046 UP000069940 UP000249989 UP000000673 UP000075880 UP000075900 UP000069272 UP000075883 UP000076408 UP000030765 UP000075884 UP000075920 UP000075886 UP000075885 UP000075902 UP000007062 UP000076407 UP000075840 UP000075881 UP000075903 UP000078541 UP000015103 UP000037069 UP000095300 UP000268350 UP000005207 UP000261640
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J005
A0A0N1INW4
A0A194Q3U1
A0A212FNW4
A0A195DI45
A0A158NQR2
+ More
A0A195BE87 A0A151WN86 E9IG54 A0A195D3P1 A0A067RTH7 A0A2J7PU34 A0A026WLG5 Q177C0 A0A1S4FCV3 E0V8V7 A0A182H7A1 W5JJ28 A0A182IZ36 A0A182RIM2 A0A2M4B849 A0A182F7I1 A0A182MBV7 A0A182Y3R4 A0A084VD37 A0A182NRV3 A0A182VZK6 A0A182QX04 A0A182PSK4 A0A182TWJ7 Q7PY15 A0A2M4A3T0 A0A182WSR7 A0A182HLM0 A0A182JRZ4 A0A182VKQ5 A0A195FSL9 T1HJQ7 A0A0L0CLP2 A0A1L8DRD4 A0A2M4CMG6 W8CCK9 A0A2M4CPJ6 A0A1I8NZR9 T1J2L3 A0A3B0JLZ1 A0A023GMF5 A0A224Z218 L7LXL1 A0A2R5LKZ7 I3KG96 I3KG97 A0A3Q3N3M1
A0A195BE87 A0A151WN86 E9IG54 A0A195D3P1 A0A067RTH7 A0A2J7PU34 A0A026WLG5 Q177C0 A0A1S4FCV3 E0V8V7 A0A182H7A1 W5JJ28 A0A182IZ36 A0A182RIM2 A0A2M4B849 A0A182F7I1 A0A182MBV7 A0A182Y3R4 A0A084VD37 A0A182NRV3 A0A182VZK6 A0A182QX04 A0A182PSK4 A0A182TWJ7 Q7PY15 A0A2M4A3T0 A0A182WSR7 A0A182HLM0 A0A182JRZ4 A0A182VKQ5 A0A195FSL9 T1HJQ7 A0A0L0CLP2 A0A1L8DRD4 A0A2M4CMG6 W8CCK9 A0A2M4CPJ6 A0A1I8NZR9 T1J2L3 A0A3B0JLZ1 A0A023GMF5 A0A224Z218 L7LXL1 A0A2R5LKZ7 I3KG96 I3KG97 A0A3Q3N3M1
PDB
2IX3
E-value=7.77488e-24,
Score=281
Ontologies
GO
PANTHER
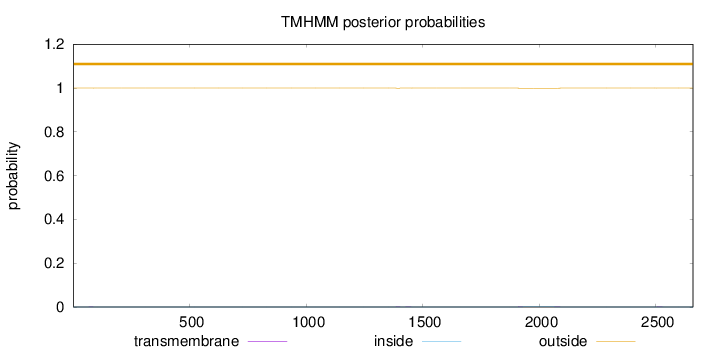
Topology
Length:
2659
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06957
Exp number, first 60 AAs:
0.00023
Total prob of N-in:
0.00024
outside
1 - 2659
Population Genetic Test Statistics
Pi
13.79778
Theta
6.344094
Tajima's D
-2.334355
CLR
181.941218
CSRT
0.00194990250487476
Interpretation
Uncertain
