Gene
KWMTBOMO06040 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002839
Annotation
PREDICTED:_protein_IWS1_homolog_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.295
Sequence
CDS
ATGGGAGGTGCAGTGAGTTCAGGGCGTGATAATAATGAGTTGATTGACAACCTGATGAGAGGCAAATACATCCGTTCCGCGGAAGTGGAGAACGTATTCCGGGCACTCGATCGTGCCGATTACATGTCTTCAGAAGTACGGGATCAGGCGTATAAGGACCTTGCCTGGAGGAATGGATCTCTGCATATGTCAGCACCGTGTATTTACAGTGAAGTAATGGAAGCCCTAGAACTGAAAACAGGCTTAACTTTCCTGAACGTCGGCTCTGGGACAGGTTACTTGAACACCTTGGTTGGTCTGATAATTGGAACCTCCGGCATAAATCACGGCATAGAAGTAAATTCTTTTGTCGTAGACTATTCCAACAAGAAACTTTCACATTTCATTGAGAACTCTCCAACTTTAGATGAATTTGACTTTTGCGAACCAAAGTTCTTTTGTGGTAACGGCTTGTGTCTGGCGCCACTTCAGTCCACGTACGATCGTGTGTACTGCGGGGCCGGGTGTCCCTATGAATATCAGAATTATTTCAAGCAGTTAATAAAGGTGGGTGGTATACTTGTGATGCCACTAAATGATAACTTGATCCAAGTCAAGCGAGTCTCGCCAACTGAATGGACCACCAAGAACCTGCTTCATGTTTCGTTCGCGTCCCTGAAGGTGCCAACGAAGGAGGAGAGCACCGAACACGTCAAACTTGACGAGCTGTGCCCGGTCCGGCTGCAGGCGCTGTCCCGCGGCGCCGTCCGCAACACGGTCCGCGCCGTGCTGGTGCAGCGCCGCGCCGAGCTGCGCACCGTCAGCGCCGGGCCCCCGCCGCGCGGCCGCCGCGTGCCGCGCCGCATCCGCATCCCCTTCGACGACAGCGACGCAGAGGAACTGAACAGTCTTCACGACTTGGACCGCGAGAGCGGAGCCAACGAAATGAACGCTCTGCTCAATTTAGTTCTGAGCATGGGACAGAATCGCGTGGCCGGAGCCCTCCGGTTCGACTCGATCGACGCCTCCAACACTTCCGAAGAGGAAGGGGACCGCCGGGCGAGCGGCCGGTCGGGGGGGGAGGAGGACAGCGGGGAGAACGAACAGAGGGCGGAGGGCAATGAACAAGGAGATGAGCCGCCAGCGGAGGGGCAGCCGTCGCAGCTCGAGCTGTGCCGGGAAATTGTCATGTTCCAGTTCCGCGGCTCCCGGGCCAGGGCGCCGGCTGTAGATTCGACTCAAGCTCGCCGATCATCCAGCCGCTCCAACTCCAGCGACAGAGCGTCAGACTCCGGCGGAAACAACGAGAACGATGCCGGGAACCAGAAACCCAACCCTAGTCCAGAGAAAACGAAGGAGTCCAAATTCAAGAAGCTCCGAACCGGGAGCGGGACCTCGGACGCGGAGCCCGAGCCGGGGACGTCCCAGACCGTCCAGACTGAGAGCGCAAAGACTGAAGCTGCTAAAACTGATGAAGAGAGCAGCACGAAACAGGAAAAAATTGAAAATTCGGTTTCGCATTCGAGCGATGCAGACGAAAGCCTGGTGAAGGATTCCCTTAGCACTGAGATGGAGTGGGAGGAGGGGGGTAAAGATGGGTCCAGCGATGAAGAAGACACTTCGAAGGACTTCGACAAGAAACGACAGAAACTGGACAGCGGAATAGGAGAGGAGAACTCGCCCGGGGGATCGTCCCCGGAGAAGACTGAGCCCAGGAGCGACCACTCCGATGCTGTGCAGTCCGACGGGAGCCTCGAGGAAGACGATGATCATCAACGAGCGCGACCTCGCAGACCCAAGCGCAGCCATCGCGACGTAGAAACTAGGTCGTCGTCTTCGTCCTCGTCCTCCGAGGAAGAGATCCCGCGGAAGGTGGACCGCGCCACGAAGAAGAAGCGTCGCGCGTGCCACGCCGCGCTGGACGTGCGGCGGCTGCGGCTGTCGCTGCTGCTGCGGCGCGAGCTGTACGCGCTGCCGCTGCCGCCCGCGCTGCGCCACTACGTCAACTACGGCCGCACGCCGCCCGCATGA
Protein
MGGAVSSGRDNNELIDNLMRGKYIRSAEVENVFRALDRADYMSSEVRDQAYKDLAWRNGSLHMSAPCIYSEVMEALELKTGLTFLNVGSGTGYLNTLVGLIIGTSGINHGIEVNSFVVDYSNKKLSHFIENSPTLDEFDFCEPKFFCGNGLCLAPLQSTYDRVYCGAGCPYEYQNYFKQLIKVGGILVMPLNDNLIQVKRVSPTEWTTKNLLHVSFASLKVPTKEESTEHVKLDELCPVRLQALSRGAVRNTVRAVLVQRRAELRTVSAGPPPRGRRVPRRIRIPFDDSDAEELNSLHDLDRESGANEMNALLNLVLSMGQNRVAGALRFDSIDASNTSEEEGDRRASGRSGGEEDSGENEQRAEGNEQGDEPPAEGQPSQLELCREIVMFQFRGSRARAPAVDSTQARRSSSRSNSSDRASDSGGNNENDAGNQKPNPSPEKTKESKFKKLRTGSGTSDAEPEPGTSQTVQTESAKTEAAKTDEESSTKQEKIENSVSHSSDADESLVKDSLSTEMEWEEGGKDGSSDEEDTSKDFDKKRQKLDSGIGEENSPGGSSPEKTEPRSDHSDAVQSDGSLEEDDDHQRARPRRPKRSHRDVETRSSSSSSSSEEEIPRKVDRATKKKRRACHAALDVRRLRLSLLLRRELYALPLPPALRHYVNYGRTPPA
Summary
Uniprot
Pubmed
EMBL
Proteomes
PRIDE
ProteinModelPortal
PDB
1KR5
E-value=4.93164e-09,
Score=148
Ontologies
GO
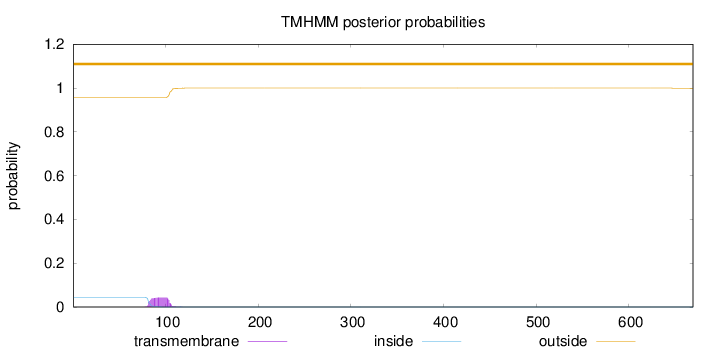
Topology
Length:
669
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.97617
Exp number, first 60 AAs:
0.00036
Total prob of N-in:
0.04327
outside
1 - 669
Population Genetic Test Statistics
Pi
19.699871
Theta
16.595014
Tajima's D
0
CLR
29.687602
CSRT
0.380330983450827
Interpretation
Uncertain
