Gene
KWMTBOMO06035
Pre Gene Modal
BGIBMGA002835
Annotation
PREDICTED:_GTP-binding_protein_10_homolog_[Amyelois_transitella]
Full name
GTP-binding protein 10 homolog
Location in the cell
Mitochondrial Reliability : 2.577
Sequence
CDS
ATGGTGCTCATATTTAGATCTCTGTTTTGTAAAGAATTAAAAAAACCACGCAATTATCTACGGTCCAAATTTATCGATTCTGTTAGACTTCATGTAAAAGGTGGAACTGGAGGCACCGGCCTTCCCAAACTGGGGGGTTTGGGAGGACAAGGAGGATGCGTTTACTGTGTTGGAACAGAAACTGCGAACCTACGTCGTATCATGAGTAATTTCAGAGGTAAAACAATCACCGCCGGTCATGGGGAAGATAGTCGTAAAACGAAAATTATTGGTACTCCGGGTGAGGATATCAAACTCGAAGTGCCTCTGGGTGTGACCGTGTATAGAGAAGATGGCACAGTCCTGGGCTCTATCGATAAAGAGGACCAAACTGTAATTATTGCGAGAGGAGGGCCAGGAGGAACTCCACAAAACAACTTCCTAGGACATTTTGGTCAAACTCATCATATACGTTTAGACCTAAAATTGATTGCTGATATTGGTCTTGTAGGTTTTCCTAATGCTGGCAAAAGTTCCTTACTAAAAGCCATTTCCAGAGCCAAGCCCAGAATAGCTAATTATCCATTCACAACCATAAAACCAAATAAAGGTGTCATGGTATATGCAGATAAACGTCAGATATCTATCGCGGACTTGCCAGGACTGATTGAAGGTGCTCATGTGAATATTGGATTGGGTCACACATTTTTAAAACATGTAGAAAGAACCAAATTGTTACTCTTTATAGCTGATGTGAAAGGTTTTCAGTTGAGCCCTAAACATCCAAATCGTTCATGCTTAGAAACAGTTCTGCTGTTAAATAAGGAACTGGAATTATACAATAAGGAGTTTTTAAATAAACCAGCAATACTTGCCATTAATAAAATGGATCTCCCTGGCTCTGAAGAAACATTACACAGGATAAAAGAGTCATACAATGATAAGGATAATATTTTAAACTCCATTCCTGAGGAGATAAGACCTGAGAGATTGATTGAATTTGATGATATCATTGGCATTTCAGCACTAGAAAGAGGAGAAAGTATAGAACAGTTGAAACAATTATTAAGAAGAAGGTTGGATCAATTTGCAGATGAGAATGACCCTGATATTGTAGAACCAAGCAAGTTACTTAGAAAACACACTAGTATGTTAAGAGAAAGAGGACCTAGAGTTACTTAG
Protein
MVLIFRSLFCKELKKPRNYLRSKFIDSVRLHVKGGTGGTGLPKLGGLGGQGGCVYCVGTETANLRRIMSNFRGKTITAGHGEDSRKTKIIGTPGEDIKLEVPLGVTVYREDGTVLGSIDKEDQTVIIARGGPGGTPQNNFLGHFGQTHHIRLDLKLIADIGLVGFPNAGKSSLLKAISRAKPRIANYPFTTIKPNKGVMVYADKRQISIADLPGLIEGAHVNIGLGHTFLKHVERTKLLLFIADVKGFQLSPKHPNRSCLETVLLLNKELELYNKEFLNKPAILAINKMDLPGSEETLHRIKESYNDKDNILNSIPEEIRPERLIEFDDIIGISALERGESIEQLKQLLRRRLDQFADENDPDIVEPSKLLRKHTSMLRERGPRVT
Summary
Similarity
Belongs to the TRAFAC class OBG-HflX-like GTPase superfamily. OBG GTPase family.
Keywords
Complete proteome
GTP-binding
Nucleotide-binding
Nucleus
Reference proteome
Ribosome biogenesis
Feature
chain GTP-binding protein 10 homolog
Uniprot
H9J000
A0A2A4J3Y8
A0A0L7L028
A0A2H1VXB9
A0A0N1PFT4
A0A194Q4C3
+ More
A0A212FNU4 S4PXW9 A0A1W4WNA7 A0A195DCH9 F4WM43 A0A195FU88 A0A158P3W7 A0A0J7L0M7 E2APZ1 A0A195BD95 A0A3L8DGM7 A0A151XA70 A0A2S2R3Y2 A0A1B6CW52 A0A195C612 A0A2S2NLK6 D6WLE5 R7TXN3 B4KIA7 B4LVH2 A0A034WIG9 A0A067QV54 B4MW97 A0A0N0BB81 A0A1B0DMU0 A0A1I8NF80 A0A2M4ASQ5 W8C5D6 A0A1B0G0D0 A0A1W4V6S1 B3NL36 A0A0A9Z9B9 A0A146MCX3 B4I5Z6 W5JK54 L7M7T7 J9JRA5 A0A1B6ED58 A0A182F1Y2 A0A1B0CSI3 A0A3B0JV20 A0A0K8U6U7 A0A1A9Y550 A0A1B0BZT7 L7M6D9 A0A1W4VKB0 A0A0M4EC40 A0A1D1VEM8 J3JV18 A0A2A3E465 A0A0A1XLC0 A0A182H2C2 Q17BY4 T1FNL0 A0A131XFW2 A0A1Y1MAB3 A0A1A9V8C0 A0A224YV79 A0A131YNH2 A0A0C9RRQ1 A0A232FFX2 G3ML75 Q9I7M2 A0A0L0CNB8 B7FNI3 A0A2J7R9C5 A0A1A9WKB5 B4PB27 T1J6A8 R4G7U3 A0A1S3D8B6 A0A1S3D8J6 B4GKW0 Q29K06 B4JR70 A0A088ARQ0 A0A1B6H227 B4QA26 A0A182UFA1 A0A210QQN5 A0A1A9ZNZ3 A0A1E1X9U7 A0A182P8M1 A0A2M4BRY0 A0A182MQ97 A0A182L0Q3 A0A3F2YSU8 E2BX46 A0A2M4BRX5 A0A023F7T7 A0A3R7PNG8 B3ML64 A0A1A8FEY0 A0A0L7RDH8
A0A212FNU4 S4PXW9 A0A1W4WNA7 A0A195DCH9 F4WM43 A0A195FU88 A0A158P3W7 A0A0J7L0M7 E2APZ1 A0A195BD95 A0A3L8DGM7 A0A151XA70 A0A2S2R3Y2 A0A1B6CW52 A0A195C612 A0A2S2NLK6 D6WLE5 R7TXN3 B4KIA7 B4LVH2 A0A034WIG9 A0A067QV54 B4MW97 A0A0N0BB81 A0A1B0DMU0 A0A1I8NF80 A0A2M4ASQ5 W8C5D6 A0A1B0G0D0 A0A1W4V6S1 B3NL36 A0A0A9Z9B9 A0A146MCX3 B4I5Z6 W5JK54 L7M7T7 J9JRA5 A0A1B6ED58 A0A182F1Y2 A0A1B0CSI3 A0A3B0JV20 A0A0K8U6U7 A0A1A9Y550 A0A1B0BZT7 L7M6D9 A0A1W4VKB0 A0A0M4EC40 A0A1D1VEM8 J3JV18 A0A2A3E465 A0A0A1XLC0 A0A182H2C2 Q17BY4 T1FNL0 A0A131XFW2 A0A1Y1MAB3 A0A1A9V8C0 A0A224YV79 A0A131YNH2 A0A0C9RRQ1 A0A232FFX2 G3ML75 Q9I7M2 A0A0L0CNB8 B7FNI3 A0A2J7R9C5 A0A1A9WKB5 B4PB27 T1J6A8 R4G7U3 A0A1S3D8B6 A0A1S3D8J6 B4GKW0 Q29K06 B4JR70 A0A088ARQ0 A0A1B6H227 B4QA26 A0A182UFA1 A0A210QQN5 A0A1A9ZNZ3 A0A1E1X9U7 A0A182P8M1 A0A2M4BRY0 A0A182MQ97 A0A182L0Q3 A0A3F2YSU8 E2BX46 A0A2M4BRX5 A0A023F7T7 A0A3R7PNG8 B3ML64 A0A1A8FEY0 A0A0L7RDH8
Pubmed
19121390
26227816
26354079
22118469
23622113
21719571
+ More
21347285 20798317 30249741 18362917 19820115 23254933 17994087 25348373 24845553 25315136 24495485 25401762 26823975 20920257 23761445 25576852 27649274 22516182 23537049 25830018 26483478 17510324 28049606 28004739 28797301 26830274 28648823 22216098 10731132 12537572 12537569 26108605 17550304 15632085 22936249 28812685 28503490 20966253 25474469
21347285 20798317 30249741 18362917 19820115 23254933 17994087 25348373 24845553 25315136 24495485 25401762 26823975 20920257 23761445 25576852 27649274 22516182 23537049 25830018 26483478 17510324 28049606 28004739 28797301 26830274 28648823 22216098 10731132 12537572 12537569 26108605 17550304 15632085 22936249 28812685 28503490 20966253 25474469
EMBL
BABH01018865
NWSH01003313
PCG66579.1
JTDY01004063
KOB68646.1
ODYU01005004
+ More
SOQ45458.1 KQ461186 KPJ07368.1 KQ459472 KPJ00199.1 AGBW02004392 OWR55414.1 GAIX01004308 JAA88252.1 KQ980989 KYN10596.1 GL888217 EGI64687.1 KQ981264 KYN44023.1 ADTU01001343 LBMM01001509 KMQ96176.1 GL441645 EFN64478.1 KQ976519 KYM82160.1 QOIP01000008 RLU19466.1 KQ982353 KYQ57210.1 GGMS01015472 MBY84675.1 GEDC01019588 GEDC01002815 JAS17710.1 JAS34483.1 KQ978220 KYM96299.1 GGMR01005425 MBY18044.1 KQ971343 EFA04101.2 AMQN01002299 KB308885 ELT96206.1 CH933807 EDW13404.1 CH940649 EDW63351.1 GAKP01004479 GAKP01004478 JAC54474.1 KK852911 KDR13913.1 CH963857 EDW75967.1 KQ438551 KOX67161.1 AJVK01007139 GGFK01010493 MBW43814.1 GAMC01004934 JAC01622.1 CCAG010014191 CH954179 EDV54614.1 GBHO01001737 GBRD01011339 GDHC01019545 JAG41867.1 JAG54485.1 JAP99083.1 GDHC01013754 GDHC01001036 JAQ04875.1 JAQ17593.1 CH480822 EDW55802.1 ADMH02001270 ETN63275.1 GACK01004673 JAA60361.1 ABLF02009653 GEDC01001419 JAS35879.1 AJWK01026095 OUUW01000010 SPP85934.1 GDHF01029932 JAI22382.1 JXJN01023293 GACK01005439 JAA59595.1 CP012523 ALC40276.1 BDGG01000005 GAU99360.1 APGK01046725 APGK01046726 BT127084 KB741067 KB631663 AEE62046.1 ENN74408.1 ERL85131.1 KZ288427 PBC25871.1 GBXI01002607 JAD11685.1 JXUM01023802 KQ560671 KXJ81345.1 CH477314 EAT43836.1 AMQM01007150 KB097572 ESN94133.1 GEFH01004045 JAP64536.1 GEZM01036651 GEZM01036650 JAV82541.1 GFPF01010352 MAA21498.1 GEDV01008577 JAP79980.1 GBYB01010051 JAG79818.1 NNAY01000271 OXU29585.1 JO842626 AEO34243.1 AE014134 AY070937 JRES01000147 KNC33815.1 BT053673 ACK77588.1 NEVH01006598 PNF37439.1 CM000158 EDW90456.1 JH431874 ACPB03010980 ACPB03010981 ACPB03010982 GAHY01002122 JAA75388.1 CH479184 EDW37276.1 CH379062 CH916372 EDV99400.1 GECZ01024568 GECZ01001023 JAS45201.1 JAS68746.1 CM000361 CM002910 EDX05556.1 KMY91079.1 NEDP02002403 OWF51029.1 GFAC01003150 JAT96038.1 GGFJ01006695 MBW55836.1 AXCM01006612 APCN01000789 GL451202 EFN79765.1 GGFJ01006696 MBW55837.1 GBBI01001407 JAC17305.1 QCYY01002197 ROT72156.1 CH902620 EDV31682.1 HAEB01010831 HAEC01006733 SBQ57358.1 KQ414614 KOC68835.1
SOQ45458.1 KQ461186 KPJ07368.1 KQ459472 KPJ00199.1 AGBW02004392 OWR55414.1 GAIX01004308 JAA88252.1 KQ980989 KYN10596.1 GL888217 EGI64687.1 KQ981264 KYN44023.1 ADTU01001343 LBMM01001509 KMQ96176.1 GL441645 EFN64478.1 KQ976519 KYM82160.1 QOIP01000008 RLU19466.1 KQ982353 KYQ57210.1 GGMS01015472 MBY84675.1 GEDC01019588 GEDC01002815 JAS17710.1 JAS34483.1 KQ978220 KYM96299.1 GGMR01005425 MBY18044.1 KQ971343 EFA04101.2 AMQN01002299 KB308885 ELT96206.1 CH933807 EDW13404.1 CH940649 EDW63351.1 GAKP01004479 GAKP01004478 JAC54474.1 KK852911 KDR13913.1 CH963857 EDW75967.1 KQ438551 KOX67161.1 AJVK01007139 GGFK01010493 MBW43814.1 GAMC01004934 JAC01622.1 CCAG010014191 CH954179 EDV54614.1 GBHO01001737 GBRD01011339 GDHC01019545 JAG41867.1 JAG54485.1 JAP99083.1 GDHC01013754 GDHC01001036 JAQ04875.1 JAQ17593.1 CH480822 EDW55802.1 ADMH02001270 ETN63275.1 GACK01004673 JAA60361.1 ABLF02009653 GEDC01001419 JAS35879.1 AJWK01026095 OUUW01000010 SPP85934.1 GDHF01029932 JAI22382.1 JXJN01023293 GACK01005439 JAA59595.1 CP012523 ALC40276.1 BDGG01000005 GAU99360.1 APGK01046725 APGK01046726 BT127084 KB741067 KB631663 AEE62046.1 ENN74408.1 ERL85131.1 KZ288427 PBC25871.1 GBXI01002607 JAD11685.1 JXUM01023802 KQ560671 KXJ81345.1 CH477314 EAT43836.1 AMQM01007150 KB097572 ESN94133.1 GEFH01004045 JAP64536.1 GEZM01036651 GEZM01036650 JAV82541.1 GFPF01010352 MAA21498.1 GEDV01008577 JAP79980.1 GBYB01010051 JAG79818.1 NNAY01000271 OXU29585.1 JO842626 AEO34243.1 AE014134 AY070937 JRES01000147 KNC33815.1 BT053673 ACK77588.1 NEVH01006598 PNF37439.1 CM000158 EDW90456.1 JH431874 ACPB03010980 ACPB03010981 ACPB03010982 GAHY01002122 JAA75388.1 CH479184 EDW37276.1 CH379062 CH916372 EDV99400.1 GECZ01024568 GECZ01001023 JAS45201.1 JAS68746.1 CM000361 CM002910 EDX05556.1 KMY91079.1 NEDP02002403 OWF51029.1 GFAC01003150 JAT96038.1 GGFJ01006695 MBW55836.1 AXCM01006612 APCN01000789 GL451202 EFN79765.1 GGFJ01006696 MBW55837.1 GBBI01001407 JAC17305.1 QCYY01002197 ROT72156.1 CH902620 EDV31682.1 HAEB01010831 HAEC01006733 SBQ57358.1 KQ414614 KOC68835.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000192223 UP000078492 UP000007755 UP000078541 UP000005205 UP000036403 UP000000311 UP000078540 UP000279307 UP000075809 UP000078542 UP000007266 UP000014760 UP000009192 UP000008792 UP000027135 UP000007798 UP000053105 UP000092462 UP000095301 UP000092444 UP000192221 UP000008711 UP000001292 UP000000673 UP000007819 UP000069272 UP000092461 UP000268350 UP000092443 UP000092460 UP000092553 UP000186922 UP000019118 UP000030742 UP000242457 UP000069940 UP000249989 UP000008820 UP000015101 UP000078200 UP000215335 UP000000803 UP000037069 UP000235965 UP000091820 UP000002282 UP000015103 UP000079169 UP000008744 UP000001819 UP000001070 UP000005203 UP000000304 UP000075902 UP000242188 UP000092445 UP000075885 UP000075883 UP000075882 UP000075840 UP000008237 UP000283509 UP000007801 UP000053825
UP000192223 UP000078492 UP000007755 UP000078541 UP000005205 UP000036403 UP000000311 UP000078540 UP000279307 UP000075809 UP000078542 UP000007266 UP000014760 UP000009192 UP000008792 UP000027135 UP000007798 UP000053105 UP000092462 UP000095301 UP000092444 UP000192221 UP000008711 UP000001292 UP000000673 UP000007819 UP000069272 UP000092461 UP000268350 UP000092443 UP000092460 UP000092553 UP000186922 UP000019118 UP000030742 UP000242457 UP000069940 UP000249989 UP000008820 UP000015101 UP000078200 UP000215335 UP000000803 UP000037069 UP000235965 UP000091820 UP000002282 UP000015103 UP000079169 UP000008744 UP000001819 UP000001070 UP000005203 UP000000304 UP000075902 UP000242188 UP000092445 UP000075885 UP000075883 UP000075882 UP000075840 UP000008237 UP000283509 UP000007801 UP000053825
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J000
A0A2A4J3Y8
A0A0L7L028
A0A2H1VXB9
A0A0N1PFT4
A0A194Q4C3
+ More
A0A212FNU4 S4PXW9 A0A1W4WNA7 A0A195DCH9 F4WM43 A0A195FU88 A0A158P3W7 A0A0J7L0M7 E2APZ1 A0A195BD95 A0A3L8DGM7 A0A151XA70 A0A2S2R3Y2 A0A1B6CW52 A0A195C612 A0A2S2NLK6 D6WLE5 R7TXN3 B4KIA7 B4LVH2 A0A034WIG9 A0A067QV54 B4MW97 A0A0N0BB81 A0A1B0DMU0 A0A1I8NF80 A0A2M4ASQ5 W8C5D6 A0A1B0G0D0 A0A1W4V6S1 B3NL36 A0A0A9Z9B9 A0A146MCX3 B4I5Z6 W5JK54 L7M7T7 J9JRA5 A0A1B6ED58 A0A182F1Y2 A0A1B0CSI3 A0A3B0JV20 A0A0K8U6U7 A0A1A9Y550 A0A1B0BZT7 L7M6D9 A0A1W4VKB0 A0A0M4EC40 A0A1D1VEM8 J3JV18 A0A2A3E465 A0A0A1XLC0 A0A182H2C2 Q17BY4 T1FNL0 A0A131XFW2 A0A1Y1MAB3 A0A1A9V8C0 A0A224YV79 A0A131YNH2 A0A0C9RRQ1 A0A232FFX2 G3ML75 Q9I7M2 A0A0L0CNB8 B7FNI3 A0A2J7R9C5 A0A1A9WKB5 B4PB27 T1J6A8 R4G7U3 A0A1S3D8B6 A0A1S3D8J6 B4GKW0 Q29K06 B4JR70 A0A088ARQ0 A0A1B6H227 B4QA26 A0A182UFA1 A0A210QQN5 A0A1A9ZNZ3 A0A1E1X9U7 A0A182P8M1 A0A2M4BRY0 A0A182MQ97 A0A182L0Q3 A0A3F2YSU8 E2BX46 A0A2M4BRX5 A0A023F7T7 A0A3R7PNG8 B3ML64 A0A1A8FEY0 A0A0L7RDH8
A0A212FNU4 S4PXW9 A0A1W4WNA7 A0A195DCH9 F4WM43 A0A195FU88 A0A158P3W7 A0A0J7L0M7 E2APZ1 A0A195BD95 A0A3L8DGM7 A0A151XA70 A0A2S2R3Y2 A0A1B6CW52 A0A195C612 A0A2S2NLK6 D6WLE5 R7TXN3 B4KIA7 B4LVH2 A0A034WIG9 A0A067QV54 B4MW97 A0A0N0BB81 A0A1B0DMU0 A0A1I8NF80 A0A2M4ASQ5 W8C5D6 A0A1B0G0D0 A0A1W4V6S1 B3NL36 A0A0A9Z9B9 A0A146MCX3 B4I5Z6 W5JK54 L7M7T7 J9JRA5 A0A1B6ED58 A0A182F1Y2 A0A1B0CSI3 A0A3B0JV20 A0A0K8U6U7 A0A1A9Y550 A0A1B0BZT7 L7M6D9 A0A1W4VKB0 A0A0M4EC40 A0A1D1VEM8 J3JV18 A0A2A3E465 A0A0A1XLC0 A0A182H2C2 Q17BY4 T1FNL0 A0A131XFW2 A0A1Y1MAB3 A0A1A9V8C0 A0A224YV79 A0A131YNH2 A0A0C9RRQ1 A0A232FFX2 G3ML75 Q9I7M2 A0A0L0CNB8 B7FNI3 A0A2J7R9C5 A0A1A9WKB5 B4PB27 T1J6A8 R4G7U3 A0A1S3D8B6 A0A1S3D8J6 B4GKW0 Q29K06 B4JR70 A0A088ARQ0 A0A1B6H227 B4QA26 A0A182UFA1 A0A210QQN5 A0A1A9ZNZ3 A0A1E1X9U7 A0A182P8M1 A0A2M4BRY0 A0A182MQ97 A0A182L0Q3 A0A3F2YSU8 E2BX46 A0A2M4BRX5 A0A023F7T7 A0A3R7PNG8 B3ML64 A0A1A8FEY0 A0A0L7RDH8
PDB
1UDX
E-value=5.22282e-36,
Score=378
Ontologies
GO
Topology
Subcellular location
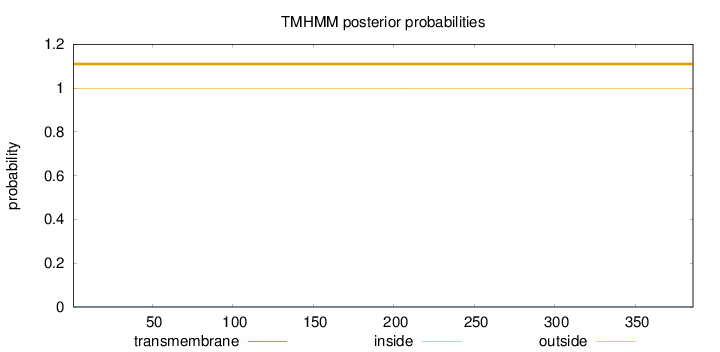
Length:
386
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00706
Exp number, first 60 AAs:
0.00285
Total prob of N-in:
0.00214
outside
1 - 386
Population Genetic Test Statistics
Pi
13.142414
Theta
10.909708
Tajima's D
0.613832
CLR
0.85477
CSRT
0.561021948902555
Interpretation
Uncertain
