Gene
KWMTBOMO06034
Annotation
PREDICTED:_post-GPI_attachment_to_proteins_factor_3_isoform_X2_[Bombyx_mori]
Full name
Post-GPI attachment to proteins factor 3
Location in the cell
PlasmaMembrane Reliability : 4.752
Sequence
CDS
ATGGGATCCTTAAAAATAACTTTAGTTACAGTATTTATTTTAAGTTGTAAATTGAGTCAGTTATCGGCGAGCGAAGGCGATCGCTTGTACATATATAAAGATTGTCTCAAACGATGCATATCGAGGAATTGTGATGAAAATGGCCTTTTGTTCAGACAGAATACAACAATACAACAGGATTTCTGGTGTAGGCTCTTCAGTTGGCGATGCATAGACGAGTGCAAGTATCATTGCATGTGGTCTGCTGTAAAGAAATTAGAGAATGCTGGTCGGCAAGTAGTCAAGTTTCATGGCAAGTGGCCATTCAAACGGATTATGGGTATGCAAGAGCCCGCATCGGTATTTGCATCTCTACTCAATTTGTTAGCAAACGCATACATGTATTCCAAATTAAGGACGGAGTTTTCAATCAAATCGAGACCTATGGTGTTACTGTGGCATTTATTTGCTCTTGTGTGCATGAACGCGTGGGTATGGTCTATGGTGTTCCACACAAGGGATACTCCGTTCACAGAGTTCATGGACTACACGTGCGCCCTGTCCATGGTCATGGGGCTGTACGTTGCTGCTGTTGTGAGAATATTGCATAGTCAGAAGAAGATCGTAGCGACGCTGCTGACGGGCTCGTTGCTGTACTTCGCGACGCACGTGCGGCTGCTGGCCACCGGGGTGGTTGACTACGACTACAACATGACCGTCAACGTTATCTTCGGCGCGGTGGGTTCGTTTATCTGGCTGGTGTACGCGGGGTACCTGAGCGCGAGAGGCTCCCGCTACTCGTCGCGCATGCTGTGGTTCGTGACGTGCTCCGCGGTCGTGCTGCCGCTGGAGCTGCTGGACTTCCCGCCGGTCCGCCGCGCGTGGGACGCGCACGCCGTGTGGCACCTCAGCACCGCGCCGCTGCCCCTGCTCTTCTACCAATTCGTGATTGAAGATCTCCGTTATACGCAGAGCAGACAAGACAAACTTTTGTAA
Protein
MGSLKITLVTVFILSCKLSQLSASEGDRLYIYKDCLKRCISRNCDENGLLFRQNTTIQQDFWCRLFSWRCIDECKYHCMWSAVKKLENAGRQVVKFHGKWPFKRIMGMQEPASVFASLLNLLANAYMYSKLRTEFSIKSRPMVLLWHLFALVCMNAWVWSMVFHTRDTPFTEFMDYTCALSMVMGLYVAAVVRILHSQKKIVATLLTGSLLYFATHVRLLATGVVDYDYNMTVNVIFGAVGSFIWLVYAGYLSARGSRYSSRMLWFVTCSAVVLPLELLDFPPVRRAWDAHAVWHLSTAPLPLLFYQFVIEDLRYTQSRQDKLL
Summary
Description
Involved in the lipid remodeling steps of GPI-anchor maturation.
Similarity
Belongs to the PGAP3 family.
Keywords
Alternative splicing
Complete proteome
Glycoprotein
Golgi apparatus
GPI-anchor biosynthesis
Membrane
Reference proteome
Signal
Transmembrane
Transmembrane helix
Feature
chain Post-GPI attachment to proteins factor 3
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9J029
A0A2A4J571
A0A3S2NBR6
A0A0N1INJ4
A0A194Q3T1
A0A212FNX3
+ More
S4PHV3 A0A2J7PTV8 Q17BP3 A0A182R2P6 A0A1Y1KMX7 A0A1Q3FDM9 A0A1Q3FDD9 A0A182SVA0 A0A1L8DY18 A0A182NH02 A0A182PJU8 B0X469 A0A182YNP6 A0A182WAP4 A0A182Q669 A0A084W9V9 A0A182H976 A0A182TIW7 A0A182VLM2 A0A182KW63 A0A182M7J0 A0A182HMC1 D6WZ63 A0A182FE76 A0A1A9WK28 A0A1A9ZP88 A0A182KA89 A0A1B0BR45 W5JE17 A0A0K8U627 A0A1A9VX77 W8B0W0 A0A1A9Y6B7 A0A182IMW0 A0A2M4BTN4 B4QDB0 A0A0K8UEK6 A0A0J9TUD3 B4HPW9 B4P0Z4 A0A067RI62 A0A1W4V1Y9 Q7K0P4-2 B3NB41 Q7Q7B9 B3MJ36 A0A1B0BR44 A0A182XCZ3 A0A0Q9X7X0 A0A293M884 A0A293MCE8 B4KNJ1 A0A1B6LFA3 Q292Z9 A0A1I8PIA6 A0A1A9Y6B6 B4GC69 A0A1B0CZY4 A0A3B0J0J3 A0A336LRU6 B4LK02 A0A226E3G5 A0A195FQ21 X2APN1 A0A195BVJ8 A0A154P0I0 A0A088AD91 A0A0Q9X817 A0A0N7ZB59 Q7K0P4 A0A3L8DFA3 A0A1B0GJF7 E1ZZE3 A0A1Z5L5V5 A0A151WT09 A0A1B6H762 V5H3B7 B7Q0M2 A0A1I8ML33 A0A131XUF5 V5IFK0 A0A0L0C577 A0A1E1XMH9 A0A0P4W320 A0A147B951 B4J6I8 A0A1B0GDL0 A0A0K2TKU7 A0A2S2R509 J9JJ61 A0A0M4EL04 A0A1B6E938 A0A1E1X9I5
S4PHV3 A0A2J7PTV8 Q17BP3 A0A182R2P6 A0A1Y1KMX7 A0A1Q3FDM9 A0A1Q3FDD9 A0A182SVA0 A0A1L8DY18 A0A182NH02 A0A182PJU8 B0X469 A0A182YNP6 A0A182WAP4 A0A182Q669 A0A084W9V9 A0A182H976 A0A182TIW7 A0A182VLM2 A0A182KW63 A0A182M7J0 A0A182HMC1 D6WZ63 A0A182FE76 A0A1A9WK28 A0A1A9ZP88 A0A182KA89 A0A1B0BR45 W5JE17 A0A0K8U627 A0A1A9VX77 W8B0W0 A0A1A9Y6B7 A0A182IMW0 A0A2M4BTN4 B4QDB0 A0A0K8UEK6 A0A0J9TUD3 B4HPW9 B4P0Z4 A0A067RI62 A0A1W4V1Y9 Q7K0P4-2 B3NB41 Q7Q7B9 B3MJ36 A0A1B0BR44 A0A182XCZ3 A0A0Q9X7X0 A0A293M884 A0A293MCE8 B4KNJ1 A0A1B6LFA3 Q292Z9 A0A1I8PIA6 A0A1A9Y6B6 B4GC69 A0A1B0CZY4 A0A3B0J0J3 A0A336LRU6 B4LK02 A0A226E3G5 A0A195FQ21 X2APN1 A0A195BVJ8 A0A154P0I0 A0A088AD91 A0A0Q9X817 A0A0N7ZB59 Q7K0P4 A0A3L8DFA3 A0A1B0GJF7 E1ZZE3 A0A1Z5L5V5 A0A151WT09 A0A1B6H762 V5H3B7 B7Q0M2 A0A1I8ML33 A0A131XUF5 V5IFK0 A0A0L0C577 A0A1E1XMH9 A0A0P4W320 A0A147B951 B4J6I8 A0A1B0GDL0 A0A0K2TKU7 A0A2S2R509 J9JJ61 A0A0M4EL04 A0A1B6E938 A0A1E1X9I5
Pubmed
19121390
26354079
22118469
23622113
17510324
28004739
+ More
25244985 24438588 26483478 20966253 18362917 19820115 20920257 23761445 24495485 17994087 22936249 17550304 24845553 10731132 12537572 12537569 17893096 12364791 18057021 15632085 23254933 30249741 20798317 28528879 25765539 25315136 26108605 29209593 28503490
25244985 24438588 26483478 20966253 18362917 19820115 20920257 23761445 24495485 17994087 22936249 17550304 24845553 10731132 12537572 12537569 17893096 12364791 18057021 15632085 23254933 30249741 20798317 28528879 25765539 25315136 26108605 29209593 28503490
EMBL
BABH01018868
BABH01018869
NWSH01003313
PCG66580.1
RSAL01000262
RVE43322.1
+ More
KQ461186 KPJ07369.1 KQ459472 KPJ00198.1 AGBW02004392 OWR55413.1 GAIX01005680 JAA86880.1 NEVH01021221 PNF19767.1 CH477319 EAT43670.1 GEZM01078374 JAV62752.1 GFDL01009370 JAV25675.1 GFDL01009492 JAV25553.1 GFDF01002746 JAV11338.1 DS232330 EDS40146.1 AXCN02000347 ATLV01021955 KE525326 KFB47003.1 JXUM01120192 KQ566369 KXJ70175.1 AXCM01000092 APCN01004400 KQ971372 EFA09753.2 JXJN01018905 ADMH02001764 ETN61145.1 GDHF01030210 JAI22104.1 GAMC01019629 GAMC01019625 GAMC01019615 GAMC01019609 JAB86926.1 GGFJ01007268 MBW56409.1 CM000362 EDX05897.1 GDHF01027293 JAI25021.1 CM002911 KMY91765.1 CH480816 EDW46638.1 CM000157 EDW89068.1 KK852619 KDR20100.1 AE013599 AY069668 CH954177 EDV59806.1 AAAB01008960 EAA11398.4 CH902619 EDV38130.1 CH933808 KRG04380.1 GFWV01011509 MAA36238.1 GFWV01013080 MAA37809.1 EDW08950.2 GEBQ01017614 JAT22363.1 CM000071 EAL24712.2 CH479181 EDW31387.1 AJVK01021142 OUUW01000001 SPP74177.1 UFQS01000063 UFQT01000063 SSW98893.1 SSX19279.1 CH940648 EDW61656.1 LNIX01000007 OXA51820.1 KQ981335 KYN42578.1 AMQN01000124 KQ976401 KYM92649.1 KQ434787 KZC05343.1 CH964282 KRG00248.1 GDRN01089010 JAI60736.1 QOIP01000009 RLU19064.1 AJWK01020252 AJWK01020253 GL435343 EFN73435.1 GFJQ02004174 JAW02796.1 KQ982769 KYQ50931.1 GECU01037197 JAS70509.1 GANP01013039 JAB71429.1 ABJB010048540 ABJB010933854 ABJB011009130 DS833048 EEC12394.1 GEFM01005928 JAP69868.1 GANP01008302 JAB76166.1 JRES01000975 KNC26599.1 GFAA01003138 JAU00297.1 GDRN01089009 JAI60737.1 GEIB01000650 JAR87309.1 CH916367 EDW01989.1 CCAG010011939 HACA01008921 CDW26282.1 GGMS01015239 MBY84442.1 ABLF02028352 CP012524 ALC42167.1 GEDC01002871 JAS34427.1 GFAC01003437 JAT95751.1
KQ461186 KPJ07369.1 KQ459472 KPJ00198.1 AGBW02004392 OWR55413.1 GAIX01005680 JAA86880.1 NEVH01021221 PNF19767.1 CH477319 EAT43670.1 GEZM01078374 JAV62752.1 GFDL01009370 JAV25675.1 GFDL01009492 JAV25553.1 GFDF01002746 JAV11338.1 DS232330 EDS40146.1 AXCN02000347 ATLV01021955 KE525326 KFB47003.1 JXUM01120192 KQ566369 KXJ70175.1 AXCM01000092 APCN01004400 KQ971372 EFA09753.2 JXJN01018905 ADMH02001764 ETN61145.1 GDHF01030210 JAI22104.1 GAMC01019629 GAMC01019625 GAMC01019615 GAMC01019609 JAB86926.1 GGFJ01007268 MBW56409.1 CM000362 EDX05897.1 GDHF01027293 JAI25021.1 CM002911 KMY91765.1 CH480816 EDW46638.1 CM000157 EDW89068.1 KK852619 KDR20100.1 AE013599 AY069668 CH954177 EDV59806.1 AAAB01008960 EAA11398.4 CH902619 EDV38130.1 CH933808 KRG04380.1 GFWV01011509 MAA36238.1 GFWV01013080 MAA37809.1 EDW08950.2 GEBQ01017614 JAT22363.1 CM000071 EAL24712.2 CH479181 EDW31387.1 AJVK01021142 OUUW01000001 SPP74177.1 UFQS01000063 UFQT01000063 SSW98893.1 SSX19279.1 CH940648 EDW61656.1 LNIX01000007 OXA51820.1 KQ981335 KYN42578.1 AMQN01000124 KQ976401 KYM92649.1 KQ434787 KZC05343.1 CH964282 KRG00248.1 GDRN01089010 JAI60736.1 QOIP01000009 RLU19064.1 AJWK01020252 AJWK01020253 GL435343 EFN73435.1 GFJQ02004174 JAW02796.1 KQ982769 KYQ50931.1 GECU01037197 JAS70509.1 GANP01013039 JAB71429.1 ABJB010048540 ABJB010933854 ABJB011009130 DS833048 EEC12394.1 GEFM01005928 JAP69868.1 GANP01008302 JAB76166.1 JRES01000975 KNC26599.1 GFAA01003138 JAU00297.1 GDRN01089009 JAI60737.1 GEIB01000650 JAR87309.1 CH916367 EDW01989.1 CCAG010011939 HACA01008921 CDW26282.1 GGMS01015239 MBY84442.1 ABLF02028352 CP012524 ALC42167.1 GEDC01002871 JAS34427.1 GFAC01003437 JAT95751.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000235965 UP000008820 UP000075900 UP000075901 UP000075884 UP000075885 UP000002320 UP000076408 UP000075920 UP000075886 UP000030765 UP000069940 UP000249989 UP000075902 UP000075903 UP000075882 UP000075883 UP000075840 UP000007266 UP000069272 UP000091820 UP000092445 UP000075881 UP000092460 UP000000673 UP000078200 UP000092443 UP000075880 UP000000304 UP000001292 UP000002282 UP000027135 UP000192221 UP000000803 UP000008711 UP000007062 UP000007801 UP000076407 UP000009192 UP000001819 UP000095300 UP000008744 UP000092462 UP000268350 UP000008792 UP000198287 UP000078541 UP000078540 UP000076502 UP000005203 UP000007798 UP000279307 UP000092461 UP000000311 UP000075809 UP000001555 UP000095301 UP000037069 UP000001070 UP000092444 UP000007819 UP000092553
UP000235965 UP000008820 UP000075900 UP000075901 UP000075884 UP000075885 UP000002320 UP000076408 UP000075920 UP000075886 UP000030765 UP000069940 UP000249989 UP000075902 UP000075903 UP000075882 UP000075883 UP000075840 UP000007266 UP000069272 UP000091820 UP000092445 UP000075881 UP000092460 UP000000673 UP000078200 UP000092443 UP000075880 UP000000304 UP000001292 UP000002282 UP000027135 UP000192221 UP000000803 UP000008711 UP000007062 UP000007801 UP000076407 UP000009192 UP000001819 UP000095300 UP000008744 UP000092462 UP000268350 UP000008792 UP000198287 UP000078541 UP000078540 UP000076502 UP000005203 UP000007798 UP000279307 UP000092461 UP000000311 UP000075809 UP000001555 UP000095301 UP000037069 UP000001070 UP000092444 UP000007819 UP000092553
Interpro
SUPFAM
SSF50891
SSF50891
Gene 3D
ProteinModelPortal
H9J029
A0A2A4J571
A0A3S2NBR6
A0A0N1INJ4
A0A194Q3T1
A0A212FNX3
+ More
S4PHV3 A0A2J7PTV8 Q17BP3 A0A182R2P6 A0A1Y1KMX7 A0A1Q3FDM9 A0A1Q3FDD9 A0A182SVA0 A0A1L8DY18 A0A182NH02 A0A182PJU8 B0X469 A0A182YNP6 A0A182WAP4 A0A182Q669 A0A084W9V9 A0A182H976 A0A182TIW7 A0A182VLM2 A0A182KW63 A0A182M7J0 A0A182HMC1 D6WZ63 A0A182FE76 A0A1A9WK28 A0A1A9ZP88 A0A182KA89 A0A1B0BR45 W5JE17 A0A0K8U627 A0A1A9VX77 W8B0W0 A0A1A9Y6B7 A0A182IMW0 A0A2M4BTN4 B4QDB0 A0A0K8UEK6 A0A0J9TUD3 B4HPW9 B4P0Z4 A0A067RI62 A0A1W4V1Y9 Q7K0P4-2 B3NB41 Q7Q7B9 B3MJ36 A0A1B0BR44 A0A182XCZ3 A0A0Q9X7X0 A0A293M884 A0A293MCE8 B4KNJ1 A0A1B6LFA3 Q292Z9 A0A1I8PIA6 A0A1A9Y6B6 B4GC69 A0A1B0CZY4 A0A3B0J0J3 A0A336LRU6 B4LK02 A0A226E3G5 A0A195FQ21 X2APN1 A0A195BVJ8 A0A154P0I0 A0A088AD91 A0A0Q9X817 A0A0N7ZB59 Q7K0P4 A0A3L8DFA3 A0A1B0GJF7 E1ZZE3 A0A1Z5L5V5 A0A151WT09 A0A1B6H762 V5H3B7 B7Q0M2 A0A1I8ML33 A0A131XUF5 V5IFK0 A0A0L0C577 A0A1E1XMH9 A0A0P4W320 A0A147B951 B4J6I8 A0A1B0GDL0 A0A0K2TKU7 A0A2S2R509 J9JJ61 A0A0M4EL04 A0A1B6E938 A0A1E1X9I5
S4PHV3 A0A2J7PTV8 Q17BP3 A0A182R2P6 A0A1Y1KMX7 A0A1Q3FDM9 A0A1Q3FDD9 A0A182SVA0 A0A1L8DY18 A0A182NH02 A0A182PJU8 B0X469 A0A182YNP6 A0A182WAP4 A0A182Q669 A0A084W9V9 A0A182H976 A0A182TIW7 A0A182VLM2 A0A182KW63 A0A182M7J0 A0A182HMC1 D6WZ63 A0A182FE76 A0A1A9WK28 A0A1A9ZP88 A0A182KA89 A0A1B0BR45 W5JE17 A0A0K8U627 A0A1A9VX77 W8B0W0 A0A1A9Y6B7 A0A182IMW0 A0A2M4BTN4 B4QDB0 A0A0K8UEK6 A0A0J9TUD3 B4HPW9 B4P0Z4 A0A067RI62 A0A1W4V1Y9 Q7K0P4-2 B3NB41 Q7Q7B9 B3MJ36 A0A1B0BR44 A0A182XCZ3 A0A0Q9X7X0 A0A293M884 A0A293MCE8 B4KNJ1 A0A1B6LFA3 Q292Z9 A0A1I8PIA6 A0A1A9Y6B6 B4GC69 A0A1B0CZY4 A0A3B0J0J3 A0A336LRU6 B4LK02 A0A226E3G5 A0A195FQ21 X2APN1 A0A195BVJ8 A0A154P0I0 A0A088AD91 A0A0Q9X817 A0A0N7ZB59 Q7K0P4 A0A3L8DFA3 A0A1B0GJF7 E1ZZE3 A0A1Z5L5V5 A0A151WT09 A0A1B6H762 V5H3B7 B7Q0M2 A0A1I8ML33 A0A131XUF5 V5IFK0 A0A0L0C577 A0A1E1XMH9 A0A0P4W320 A0A147B951 B4J6I8 A0A1B0GDL0 A0A0K2TKU7 A0A2S2R509 J9JJ61 A0A0M4EL04 A0A1B6E938 A0A1E1X9I5
Ontologies
PANTHER
Topology
Subcellular location
Golgi apparatus membrane
SignalP
Position: 1 - 23,
Likelihood: 0.973212
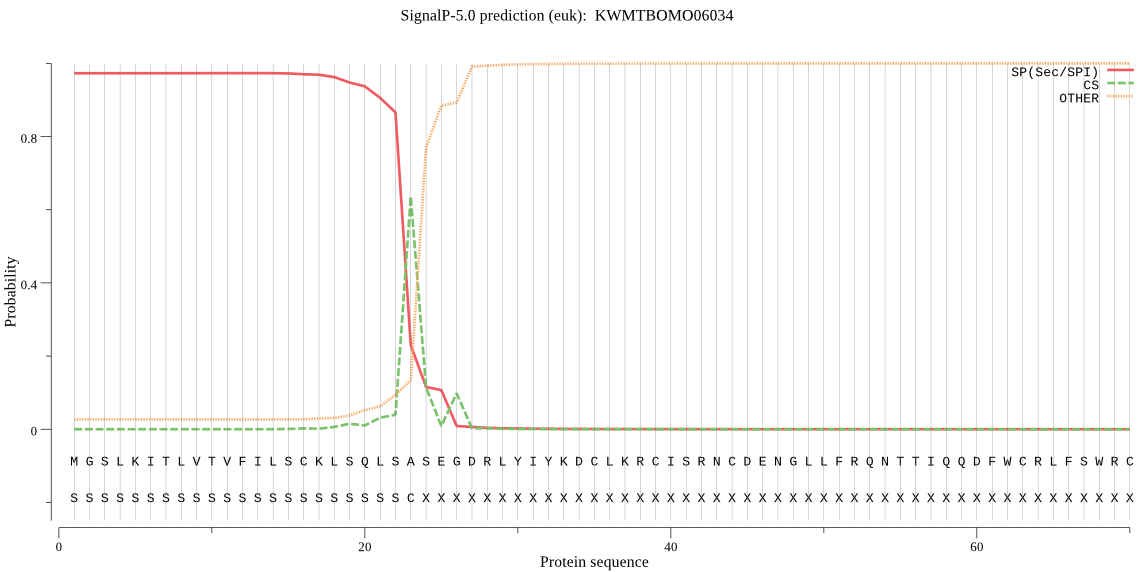
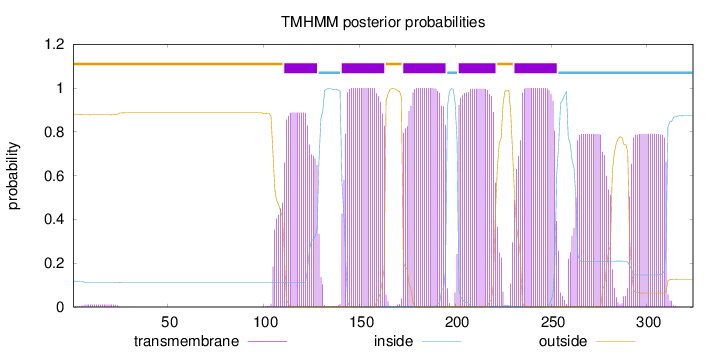
Length:
324
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
133.90092
Exp number, first 60 AAs:
0.21825
Total prob of N-in:
0.12065
outside
1 - 110
TMhelix
111 - 128
inside
129 - 140
TMhelix
141 - 163
outside
164 - 172
TMhelix
173 - 195
inside
196 - 201
TMhelix
202 - 221
outside
222 - 230
TMhelix
231 - 253
inside
254 - 324
Population Genetic Test Statistics
Pi
16.640964
Theta
15.84963
Tajima's D
0.665943
CLR
1.255663
CSRT
0.566171691415429
Interpretation
Uncertain
