Gene
KWMTBOMO06027
Pre Gene Modal
BGIBMGA002870
Annotation
fanconi_anemia_group_J_protein_[Danaus_plexippus]
Full name
Regulator of telomere elongation helicase 1 homolog
Location in the cell
Nuclear Reliability : 2.745
Sequence
CDS
ATGCCTGATATAATGATATCCGGTATTCCCGTAAGTTTTCCCTTCGACCCTTACGATGTGCAGAAAGAATACATGTGCCGTGTAATTGAGAGCCTTCAGAATAACACTAACGCAATATTGGAGTCTCCTACCGGAACTGGAAAGACCCTTAGTCTATTGTGTTCATCATTAGCATGGCTACTAGTGAAAAAGGCTCAACTGCAAATGAACGCGCAGGTCGGTAATTTTTCAGATCACAGCGGTTTTACTGGCAAGTTAATAGATAACTTAAAATCTGGCACTGGAAAATCCAAGGATGCGTCTTGGGGTATGCCTAAAATCATATATTCTTCCAGAACACATTCACAATTAACACAGGCTATGCAGGAATTGAAGAGGTCAAGTTACAAGCATGTCAAAGCTGCGGTCTTGGGCTCCCGAGATCAGATGTGCATTCATCCTGAAGTTTCGAAAGAACCAAACAACAACAATAAAGTAAATATGTGCCAACTAAAAGTGAAATCTAGAACTTGCTTCTTTTATAACAATGTTGAATCTAAAAAGGATGACAGAGCAGTGAAAGGTGATGATATTTTAGATATAGAAGATTTAGTAGGTGTAGGAAAGAAATTAAAATGCTGTCCATATTATTTATCGAAGGAGCTAAAACAAGATGCTGATATAATATTTATGCCATATAATTATTTATTGGATCCAAAATCAAGAAAGGCAAATGGAGTGGAACTCATGAATAACATTGTTATTTTGGATGAGGCTCATAATGTTGAAAAAATGTGTGAAGAATCTGCATCTCTTCAGATTAGGACCACAGATATAGCTCTGTGTATTGATGAAATAACACATGTCATGAAGAGCTTTAGTGAGAATAGTGAGGAGCAGATAGATGCAACAGTTGATAGTAACCAACCTAAAGATTTTACTTGTGATGATTTATGTATACTGAAAGAAATGATGCTGGCCTTTGAAAAGGCTGTAGATGATATAGAAGTTGGGAGAGAGGGTAGTACATATCCAGGTGGTTTTATATTTGATTTGCTGTCCAAGGCTGAAATCAGAGATCATAACCAAATGGCCGTGATAACTATGATTGAGAATCTCATTCAATATCTAGCAACAGCAAGTTCATCACCCTTTCAACGCAAAGGTGTTGGGTTGCAGAAAATTGTAGATCTTCTAAATGTTGTGTTTAGTGGCACTAGTCATTCACACAAAGAAAGGGTTAAGCTATGTTACAAAGTACATGTACAAGTTGAGGATAAAAAAAGCAATAAGAAAACAGAAGGTTGGGGTGCACTGAAGCCGAACAGTTCTAAATCTACAGAGAGGATACTAAGTTACTGGTGCTTCAGCCCTGGTTTTGGCATGAAGCAATTACTTGAACAGAATGTTAGAAGCATAATATTGACAAGTGGTACTTTAGCACCTTTAAAACCCCTCATATCTGAACTGGGTATTCCAATAGGTGTACAGTTAGAAAATCCACATATTGTTAAATCAAATCAAATATGTGTAAAAATTATTGGACAAGGGCCAGATGGGGTCCAACTAAATTCTAATTATCAGAACAGAGACAATCCTAAGTATATATCCTCTTTGGGAAGAACAATATTAAGTTTTAGCAGAGTTGTACCAGATGGACTTTTAGTATTCTTCCCTTCATACCCAATCATGAATAAATGCCAAGAAATGTGGCAGTCTGAAGGAATTTGGACTAATATTAACAGCATCAAACCAATATTTGTGGAACCGCAAAGGAAAGATACATTCAATGCTATCATTAAAGATTACTACAGCAAAATAAGTGATCCAAGTACACGGGGTGCATGTTTTATGGCAGTTTGTAGAGGTAAAGTGTCAGAAGGGTTGGATTTTGCTGATATGAATGGAAGAGCTGTGATAATAACTGGGTTACCATTTCCTCCTCTTAAGGATCCAAGAATTGTATTGAAGAAGAAATATTTAGACGAATTGAGAATGAAAGAAAAGGACTTTTTGTCAGGAGACGAATGGTACTCTTTAGAAGCAACTAGAGCCGTCAATCAGGCTATTGGCAGAGTTATAAGGCATCAGAATGATTATGGTGCTATTCTTCTGTGTGATAGCCGATTCAACAGTCCAAAATTAAAAGGTCAGTTGTCAGCTTGGCTACGTAATTACATAAATGTGTCCAATAAATTTGGTGAGACTGTCAGTGAAATATGCCGCTTCTATAAGAATGCTGAAATGGCATTGCCTGCTCCAAAACTAAAGCCGTTGATGTCAAATGAATCAAACAGACAACAATCTGTCAATGTTGGTGGAGTATCATTTCCAATTACAGTTGCTAGGACTGTTAAATCTACTAAAAAGAATAGTTTAGTGGCAATAAACAAAAGAACTGCTGAAGACAATTTATTAGCAAATGTATCTAAAAAGAAAAAATACAAAATTAAACCCTTTGGTTTTGATGAAAATTTGAAAAGTGAACCTAGTAACATAGTAGAGAAAACTGCACCTACATCACTAGTAGATTTTGTTAAAGAAATAAAGGTTTTATTACAACCAGAACAATATAAATTATTTCAAGGTTCAATATCAGCTTATAAAAGAGAAGGGGATTTTTCAGCATTTGTGACCACCCTAGGAATTTTGTTACCAAGAACTTAA
Protein
MPDIMISGIPVSFPFDPYDVQKEYMCRVIESLQNNTNAILESPTGTGKTLSLLCSSLAWLLVKKAQLQMNAQVGNFSDHSGFTGKLIDNLKSGTGKSKDASWGMPKIIYSSRTHSQLTQAMQELKRSSYKHVKAAVLGSRDQMCIHPEVSKEPNNNNKVNMCQLKVKSRTCFFYNNVESKKDDRAVKGDDILDIEDLVGVGKKLKCCPYYLSKELKQDADIIFMPYNYLLDPKSRKANGVELMNNIVILDEAHNVEKMCEESASLQIRTTDIALCIDEITHVMKSFSENSEEQIDATVDSNQPKDFTCDDLCILKEMMLAFEKAVDDIEVGREGSTYPGGFIFDLLSKAEIRDHNQMAVITMIENLIQYLATASSSPFQRKGVGLQKIVDLLNVVFSGTSHSHKERVKLCYKVHVQVEDKKSNKKTEGWGALKPNSSKSTERILSYWCFSPGFGMKQLLEQNVRSIILTSGTLAPLKPLISELGIPIGVQLENPHIVKSNQICVKIIGQGPDGVQLNSNYQNRDNPKYISSLGRTILSFSRVVPDGLLVFFPSYPIMNKCQEMWQSEGIWTNINSIKPIFVEPQRKDTFNAIIKDYYSKISDPSTRGACFMAVCRGKVSEGLDFADMNGRAVIITGLPFPPLKDPRIVLKKKYLDELRMKEKDFLSGDEWYSLEATRAVNQAIGRVIRHQNDYGAILLCDSRFNSPKLKGQLSAWLRNYINVSNKFGETVSEICRFYKNAEMALPAPKLKPLMSNESNRQQSVNVGGVSFPITVARTVKSTKKNSLVAINKRTAEDNLLANVSKKKKYKIKPFGFDENLKSEPSNIVEKTAPTSLVDFVKEIKVLLQPEQYKLFQGSISAYKREGDFSAFVTTLGILLPRT
Summary
Description
ATP-dependent DNA helicase implicated in DNA repair and the maintenance of genomic stability. Acts as an anti-recombinase to counteract toxic recombination and limit crossover during meiosis. Regulates meiotic recombination and crossover homeostasis by physically dissociating strand invasion events and thereby promotes noncrossover repair by meiotic synthesis dependent strand annealing (SDSA) as well as disassembly of D loop recombination intermediates.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Similarity
Belongs to the helicase family. RAD3/XPD subfamily.
Keywords
4Fe-4S
ATP-binding
Complete proteome
DNA damage
DNA repair
DNA-binding
Helicase
Hydrolase
Iron
Iron-sulfur
Metal-binding
Nucleotide-binding
Nucleus
Reference proteome
Phosphoprotein
Feature
chain Regulator of telomere elongation helicase 1 homolog
Uniprot
H9J035
A0A2H1WP52
A0A212FNT3
A0A0N1IBH3
A0A0L7L3L6
A0A194Q5A6
+ More
A0A0C9RZ03 A0A151X296 A0A195FIS8 A0A195E471 A0A195BT71 E2AJX8 A0A0N0BBZ8 A0A026WGY7 A0A195D325 F4W6K7 A0A158P2B2 E2C016 A0A2A3EJ04 A0A232F640 A0A154P5P8 A0A3R7MH68 E9JAI1 Q16X92 B0W9F4 A0A2K7P6B7 A0A1Q3EW05 A0A336L6I5 A0A0K8VFB4 A0A0K8VZL5 A0A310SHL9 A0A0P4WD16 A0A034W2S9 A0A0A1WV39 W8BLX8 A0A182RMV3 A0A336L6N2 A0A0L0BRH8 A0A182ME41 A0A1I8MHF9 A0A182W5F6 B3MSG8 B4JNS2 A0A1I8PPA1 A0A0M5J160 A0A0P5ZJ03 A0A0P5DSB6 A0A0P5RTG5 A0A0P6ADZ3 A0A0P5DD32 A0A0P5V4B7 A0A2J7QUE4 B4L1Z2 A0A182XXV8 A0A182KC05 B4PZB4 A0A3L8E0B1 A0A3B0J6K2 B4M891 E9GJP0 A0A0N8EPH5 U4UI39 A0A1W4VFH5 Q7QEI1 A0A0J7KNW7 A0A182TKD5 A0A1B0A073 B3NSW1 A0A182IA08 A0A1W4VT40 A0A084VQ93 A0A1W4VSF6 Q29FS3 A0A182X7D1 B4GU19 A0A182VKP5 A0A182PWK2 A0A1A9WGJ1 A0A1Y1LUN1 A0A1B0CHR4 A0A1A9XDU5 A0A1B0B8H1 A0A182JAL0 A0A182N072 A0A0P4ZVY6 A0A0P4ZI32 A0A0P4ZR51 A0A0P5STK6 A0A0P5TA30 A0A0P4ZRG7 A0A0P5TA38 B4NDG5 A0A1A9VRD2 M9PGH6 Q9W484 A0A182QAZ7 A0A182MAW6 A0A182WI58 A0A0P5SEI0 A0A1B0D175
A0A0C9RZ03 A0A151X296 A0A195FIS8 A0A195E471 A0A195BT71 E2AJX8 A0A0N0BBZ8 A0A026WGY7 A0A195D325 F4W6K7 A0A158P2B2 E2C016 A0A2A3EJ04 A0A232F640 A0A154P5P8 A0A3R7MH68 E9JAI1 Q16X92 B0W9F4 A0A2K7P6B7 A0A1Q3EW05 A0A336L6I5 A0A0K8VFB4 A0A0K8VZL5 A0A310SHL9 A0A0P4WD16 A0A034W2S9 A0A0A1WV39 W8BLX8 A0A182RMV3 A0A336L6N2 A0A0L0BRH8 A0A182ME41 A0A1I8MHF9 A0A182W5F6 B3MSG8 B4JNS2 A0A1I8PPA1 A0A0M5J160 A0A0P5ZJ03 A0A0P5DSB6 A0A0P5RTG5 A0A0P6ADZ3 A0A0P5DD32 A0A0P5V4B7 A0A2J7QUE4 B4L1Z2 A0A182XXV8 A0A182KC05 B4PZB4 A0A3L8E0B1 A0A3B0J6K2 B4M891 E9GJP0 A0A0N8EPH5 U4UI39 A0A1W4VFH5 Q7QEI1 A0A0J7KNW7 A0A182TKD5 A0A1B0A073 B3NSW1 A0A182IA08 A0A1W4VT40 A0A084VQ93 A0A1W4VSF6 Q29FS3 A0A182X7D1 B4GU19 A0A182VKP5 A0A182PWK2 A0A1A9WGJ1 A0A1Y1LUN1 A0A1B0CHR4 A0A1A9XDU5 A0A1B0B8H1 A0A182JAL0 A0A182N072 A0A0P4ZVY6 A0A0P4ZI32 A0A0P4ZR51 A0A0P5STK6 A0A0P5TA30 A0A0P4ZRG7 A0A0P5TA38 B4NDG5 A0A1A9VRD2 M9PGH6 Q9W484 A0A182QAZ7 A0A182MAW6 A0A182WI58 A0A0P5SEI0 A0A1B0D175
EC Number
3.6.4.12
Pubmed
19121390
22118469
26354079
26227816
20798317
24508170
+ More
21719571 21347285 28648823 21282665 17510324 25348373 25830018 24495485 26108605 25315136 17994087 25244985 30249741 21292972 23537049 12364791 24438588 15632085 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17989248 18327897
21719571 21347285 28648823 21282665 17510324 25348373 25830018 24495485 26108605 25315136 17994087 25244985 30249741 21292972 23537049 12364791 24438588 15632085 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17989248 18327897
EMBL
BABH01018874
ODYU01010033
SOQ54851.1
AGBW02004392
OWR55404.1
KQ461186
+ More
KPJ07377.1 JTDY01003232 KOB69911.1 KQ459472 KPJ00190.1 GBYB01014614 GBYB01014615 GBYB01014616 JAG84381.1 JAG84382.1 JAG84383.1 KQ982580 KYQ54508.1 KQ981523 KYN40303.1 KQ979657 KYN19953.1 KQ976408 KYM91136.1 GL440100 EFN66294.1 KQ435966 KOX67941.1 KK107250 EZA54344.1 KQ976885 KYN07303.1 GL887707 EGI70342.1 ADTU01007051 GL451712 EFN78712.1 KZ288227 PBC31745.1 NNAY01000911 OXU25960.1 KQ434809 KZC06664.1 QCYY01000940 ROT81658.1 GL770368 EFZ10173.1 CH477545 DS231864 GFDL01015555 JAV19490.1 UFQS01002091 UFQT01002091 SSX13190.1 SSX32629.1 GDHF01014761 JAI37553.1 GDHF01007961 JAI44353.1 KQ763209 OAD55149.1 GDRN01080265 JAI62259.1 GAKP01009071 JAC49881.1 GBXI01011791 JAD02501.1 GAMC01004460 JAC02096.1 UFQS01001166 UFQT01001166 SSX09317.1 SSX29219.1 JRES01001467 KNC22680.1 AXCM01014096 CH902622 CH916371 CP012528 ALC49144.1 GDIP01225111 GDIP01044398 LRGB01001363 JAM59317.1 KZS12158.1 GDIP01169111 JAJ54291.1 GDIQ01114576 JAL37150.1 GDIP01032041 JAM71674.1 GDIP01158072 JAJ65330.1 GDIP01104977 JAL98737.1 NEVH01011186 PNF32201.1 CH933810 CM000162 QOIP01000002 RLU25923.1 OUUW01000003 SPP77637.1 CH940653 GL732548 EFX80285.1 GDIQ01006668 JAN88069.1 KB632308 ERL92013.1 AAAB01008847 LBMM01004880 KMQ92022.1 CH954180 APCN01001081 ATLV01015151 KE525003 KFB40137.1 CH379065 CH479190 GEZM01046290 JAV77354.1 AJWK01012600 JXJN01009934 GDIP01211670 JAJ11732.1 GDIP01212516 JAJ10886.1 GDIP01209068 JAJ14334.1 GDIP01138674 JAL65040.1 GDIP01133099 JAL70615.1 GDIP01209069 JAJ14333.1 GDIP01130476 JAL73238.1 CH964239 AE014298 AGB95108.1 BT021290 EU217688 EU217689 EU217690 EU217691 EU217692 EU217693 EU217694 EU217695 EU217696 EU217697 EU217698 EU217699 AXCN02001428 AXCM01000817 GDIP01140776 JAL62938.1 AJVK01002427 AJVK01002428 AJVK01002429
KPJ07377.1 JTDY01003232 KOB69911.1 KQ459472 KPJ00190.1 GBYB01014614 GBYB01014615 GBYB01014616 JAG84381.1 JAG84382.1 JAG84383.1 KQ982580 KYQ54508.1 KQ981523 KYN40303.1 KQ979657 KYN19953.1 KQ976408 KYM91136.1 GL440100 EFN66294.1 KQ435966 KOX67941.1 KK107250 EZA54344.1 KQ976885 KYN07303.1 GL887707 EGI70342.1 ADTU01007051 GL451712 EFN78712.1 KZ288227 PBC31745.1 NNAY01000911 OXU25960.1 KQ434809 KZC06664.1 QCYY01000940 ROT81658.1 GL770368 EFZ10173.1 CH477545 DS231864 GFDL01015555 JAV19490.1 UFQS01002091 UFQT01002091 SSX13190.1 SSX32629.1 GDHF01014761 JAI37553.1 GDHF01007961 JAI44353.1 KQ763209 OAD55149.1 GDRN01080265 JAI62259.1 GAKP01009071 JAC49881.1 GBXI01011791 JAD02501.1 GAMC01004460 JAC02096.1 UFQS01001166 UFQT01001166 SSX09317.1 SSX29219.1 JRES01001467 KNC22680.1 AXCM01014096 CH902622 CH916371 CP012528 ALC49144.1 GDIP01225111 GDIP01044398 LRGB01001363 JAM59317.1 KZS12158.1 GDIP01169111 JAJ54291.1 GDIQ01114576 JAL37150.1 GDIP01032041 JAM71674.1 GDIP01158072 JAJ65330.1 GDIP01104977 JAL98737.1 NEVH01011186 PNF32201.1 CH933810 CM000162 QOIP01000002 RLU25923.1 OUUW01000003 SPP77637.1 CH940653 GL732548 EFX80285.1 GDIQ01006668 JAN88069.1 KB632308 ERL92013.1 AAAB01008847 LBMM01004880 KMQ92022.1 CH954180 APCN01001081 ATLV01015151 KE525003 KFB40137.1 CH379065 CH479190 GEZM01046290 JAV77354.1 AJWK01012600 JXJN01009934 GDIP01211670 JAJ11732.1 GDIP01212516 JAJ10886.1 GDIP01209068 JAJ14334.1 GDIP01138674 JAL65040.1 GDIP01133099 JAL70615.1 GDIP01209069 JAJ14333.1 GDIP01130476 JAL73238.1 CH964239 AE014298 AGB95108.1 BT021290 EU217688 EU217689 EU217690 EU217691 EU217692 EU217693 EU217694 EU217695 EU217696 EU217697 EU217698 EU217699 AXCN02001428 AXCM01000817 GDIP01140776 JAL62938.1 AJVK01002427 AJVK01002428 AJVK01002429
Proteomes
UP000005204
UP000007151
UP000053240
UP000037510
UP000053268
UP000075809
+ More
UP000078541 UP000078492 UP000078540 UP000000311 UP000053105 UP000053097 UP000078542 UP000007755 UP000005205 UP000008237 UP000242457 UP000215335 UP000076502 UP000283509 UP000008820 UP000002320 UP000075900 UP000037069 UP000075883 UP000095301 UP000075920 UP000007801 UP000001070 UP000095300 UP000092553 UP000076858 UP000235965 UP000009192 UP000076408 UP000075881 UP000002282 UP000279307 UP000268350 UP000008792 UP000000305 UP000030742 UP000192221 UP000007062 UP000036403 UP000075902 UP000092445 UP000008711 UP000075840 UP000030765 UP000001819 UP000076407 UP000008744 UP000075903 UP000075885 UP000091820 UP000092461 UP000092443 UP000092460 UP000075880 UP000075884 UP000007798 UP000078200 UP000000803 UP000075886 UP000092462
UP000078541 UP000078492 UP000078540 UP000000311 UP000053105 UP000053097 UP000078542 UP000007755 UP000005205 UP000008237 UP000242457 UP000215335 UP000076502 UP000283509 UP000008820 UP000002320 UP000075900 UP000037069 UP000075883 UP000095301 UP000075920 UP000007801 UP000001070 UP000095300 UP000092553 UP000076858 UP000235965 UP000009192 UP000076408 UP000075881 UP000002282 UP000279307 UP000268350 UP000008792 UP000000305 UP000030742 UP000192221 UP000007062 UP000036403 UP000075902 UP000092445 UP000008711 UP000075840 UP000030765 UP000001819 UP000076407 UP000008744 UP000075903 UP000075885 UP000091820 UP000092461 UP000092443 UP000092460 UP000075880 UP000075884 UP000007798 UP000078200 UP000000803 UP000075886 UP000092462
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9J035
A0A2H1WP52
A0A212FNT3
A0A0N1IBH3
A0A0L7L3L6
A0A194Q5A6
+ More
A0A0C9RZ03 A0A151X296 A0A195FIS8 A0A195E471 A0A195BT71 E2AJX8 A0A0N0BBZ8 A0A026WGY7 A0A195D325 F4W6K7 A0A158P2B2 E2C016 A0A2A3EJ04 A0A232F640 A0A154P5P8 A0A3R7MH68 E9JAI1 Q16X92 B0W9F4 A0A2K7P6B7 A0A1Q3EW05 A0A336L6I5 A0A0K8VFB4 A0A0K8VZL5 A0A310SHL9 A0A0P4WD16 A0A034W2S9 A0A0A1WV39 W8BLX8 A0A182RMV3 A0A336L6N2 A0A0L0BRH8 A0A182ME41 A0A1I8MHF9 A0A182W5F6 B3MSG8 B4JNS2 A0A1I8PPA1 A0A0M5J160 A0A0P5ZJ03 A0A0P5DSB6 A0A0P5RTG5 A0A0P6ADZ3 A0A0P5DD32 A0A0P5V4B7 A0A2J7QUE4 B4L1Z2 A0A182XXV8 A0A182KC05 B4PZB4 A0A3L8E0B1 A0A3B0J6K2 B4M891 E9GJP0 A0A0N8EPH5 U4UI39 A0A1W4VFH5 Q7QEI1 A0A0J7KNW7 A0A182TKD5 A0A1B0A073 B3NSW1 A0A182IA08 A0A1W4VT40 A0A084VQ93 A0A1W4VSF6 Q29FS3 A0A182X7D1 B4GU19 A0A182VKP5 A0A182PWK2 A0A1A9WGJ1 A0A1Y1LUN1 A0A1B0CHR4 A0A1A9XDU5 A0A1B0B8H1 A0A182JAL0 A0A182N072 A0A0P4ZVY6 A0A0P4ZI32 A0A0P4ZR51 A0A0P5STK6 A0A0P5TA30 A0A0P4ZRG7 A0A0P5TA38 B4NDG5 A0A1A9VRD2 M9PGH6 Q9W484 A0A182QAZ7 A0A182MAW6 A0A182WI58 A0A0P5SEI0 A0A1B0D175
A0A0C9RZ03 A0A151X296 A0A195FIS8 A0A195E471 A0A195BT71 E2AJX8 A0A0N0BBZ8 A0A026WGY7 A0A195D325 F4W6K7 A0A158P2B2 E2C016 A0A2A3EJ04 A0A232F640 A0A154P5P8 A0A3R7MH68 E9JAI1 Q16X92 B0W9F4 A0A2K7P6B7 A0A1Q3EW05 A0A336L6I5 A0A0K8VFB4 A0A0K8VZL5 A0A310SHL9 A0A0P4WD16 A0A034W2S9 A0A0A1WV39 W8BLX8 A0A182RMV3 A0A336L6N2 A0A0L0BRH8 A0A182ME41 A0A1I8MHF9 A0A182W5F6 B3MSG8 B4JNS2 A0A1I8PPA1 A0A0M5J160 A0A0P5ZJ03 A0A0P5DSB6 A0A0P5RTG5 A0A0P6ADZ3 A0A0P5DD32 A0A0P5V4B7 A0A2J7QUE4 B4L1Z2 A0A182XXV8 A0A182KC05 B4PZB4 A0A3L8E0B1 A0A3B0J6K2 B4M891 E9GJP0 A0A0N8EPH5 U4UI39 A0A1W4VFH5 Q7QEI1 A0A0J7KNW7 A0A182TKD5 A0A1B0A073 B3NSW1 A0A182IA08 A0A1W4VT40 A0A084VQ93 A0A1W4VSF6 Q29FS3 A0A182X7D1 B4GU19 A0A182VKP5 A0A182PWK2 A0A1A9WGJ1 A0A1Y1LUN1 A0A1B0CHR4 A0A1A9XDU5 A0A1B0B8H1 A0A182JAL0 A0A182N072 A0A0P4ZVY6 A0A0P4ZI32 A0A0P4ZR51 A0A0P5STK6 A0A0P5TA30 A0A0P4ZRG7 A0A0P5TA38 B4NDG5 A0A1A9VRD2 M9PGH6 Q9W484 A0A182QAZ7 A0A182MAW6 A0A182WI58 A0A0P5SEI0 A0A1B0D175
PDB
5SVA
E-value=4.93349e-58,
Score=572
Ontologies
GO
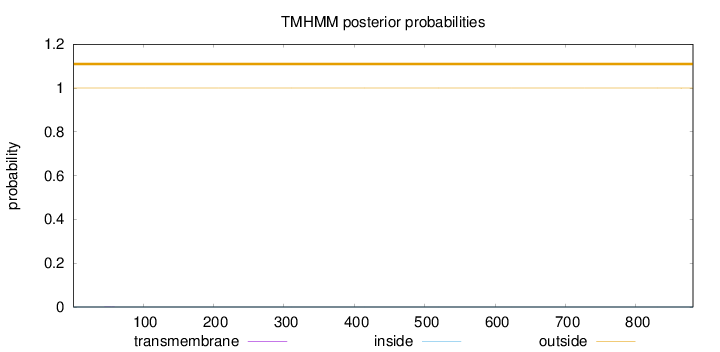
Topology
Subcellular location
Nucleus
Length:
881
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00588999999999998
Exp number, first 60 AAs:
0.00262
Total prob of N-in:
0.00014
outside
1 - 881
Population Genetic Test Statistics
Pi
12.88839
Theta
15.51874
Tajima's D
0.395327
CLR
0.067125
CSRT
0.494075296235188
Interpretation
Uncertain
