Gene
KWMTBOMO06020
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 3.099
Sequence
CDS
ATGCTGGGAGTCCTCAACAGAGCGAAGCGGTACTTCACGCCTGGACAAAGGCTTCTGCTTTATAAAGCACAAGTTCGGCCTCGCGTGGAGTACTGCTCCCATCTCTGGGCCGGGGCTCCCAAATACCAGCTTCTTCCATTTGACTCCATACAGAGGAGGGCCGTTCGGATTGTCGATAATCCCAACCTCACGGATCGTTTGGAACCTCTGGGTCTGCGGAGGGACTTCGGTTCCCTCTGTATTTTGTACCGTATGTTCCATGGGGAGTGCTCTGAGGAATTGTTCGAGATGATACCGGCATCTCGTTTTTACCATCGCATCGCCCGCCACCGGAGTAGAGTTCATCCGTACTACCTGGAGCCACTGCGGTCATCCACAGTGCGTTTCCAGAGATCTTTTTTGCCACGTACCATCCGGCTATGGAATGAGCTCCCCTCCACGGTGTTTCCCGAGCGCTATGACATGTCCTTCTTCAAACGAGGCTTGTGGAGAGTATTAAGCGGTAGGCAGCGGCTTGGCTCTGCCCCTGGCATTGCTGAAGTCCATGGGCAACGGTAA
Protein
MLGVLNRAKRYFTPGQRLLLYKAQVRPRVEYCSHLWAGAPKYQLLPFDSIQRRAVRIVDNPNLTDRLEPLGLRRDFGSLCILYRMFHGECSEELFEMIPASRFYHRIARHRSRVHPYYLEPLRSSTVRFQRSFLPRTIRLWNELPSTVFPERYDMSFFKRGLWRVLSGRQRLGSAPGIAEVHGQR
Summary
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Uniprot
Q5KTM5
Q6UV17
A0A3S2LP03
A0A3S2P9Q3
A0A3S2NHP4
A0A3S2NMS5
+ More
B9WPT5 A0A3S2NMX8 A0A3S2LQP1 A0A2H1WAR9 A0A2H1VFZ3 J9GT67 T2M6M7 A0A3S2TFH4 A0A3S2PF93 A0A2H1V2F4 A0A3D5S1K1 A0A3S2PHM8 A0A1Y1S3P2 A0A0P4VXU0 A0A3S2LI48 A0A2H1VUV7 A0A2H1WY65 A0A2H1WCG9 A0A2H1W7F5 A0A2H1VQC3 A0A2G8JWN9 W4Z5Z2 A0A1E1XGQ7 A0A131XCJ7 A0A1E1XPS6 A0A2G8KRG4 W4XMI9 A0A0C9SB62 A0A131Y4D1 A0A147BJ69 A0A1E1XVB3 A0A1E1XN74 A0A1E1XNW6 A0A0K8RG16 A0A2I0UC10 A0A1E1XGZ3 A0A2H9T4Q5 A0A131Y4F6 A0A1E1X2P5 A0A016UDM4 A0A131Y8X0 A0A147BJQ0 A0A016UEC4 A0A3B3RE95 A0A131Y3E3 A0A131Y3W4 W4YZP0 A0A131Y3C2 V5H708 A0A147BLD9 A0A131XRE4 K7EWV4 A0A2I0UP23 A0A3B3RWV8 W4Y8J8 W4ZK03 A0A091HUB5 A0A131Y4Q4 A0A1E1XND6 A0A2I0UDR0 W4ZIH4 K7EYK2 A0A091I8M3 K7EYD9 K7EY85 K7EYG1
B9WPT5 A0A3S2NMX8 A0A3S2LQP1 A0A2H1WAR9 A0A2H1VFZ3 J9GT67 T2M6M7 A0A3S2TFH4 A0A3S2PF93 A0A2H1V2F4 A0A3D5S1K1 A0A3S2PHM8 A0A1Y1S3P2 A0A0P4VXU0 A0A3S2LI48 A0A2H1VUV7 A0A2H1WY65 A0A2H1WCG9 A0A2H1W7F5 A0A2H1VQC3 A0A2G8JWN9 W4Z5Z2 A0A1E1XGQ7 A0A131XCJ7 A0A1E1XPS6 A0A2G8KRG4 W4XMI9 A0A0C9SB62 A0A131Y4D1 A0A147BJ69 A0A1E1XVB3 A0A1E1XN74 A0A1E1XNW6 A0A0K8RG16 A0A2I0UC10 A0A1E1XGZ3 A0A2H9T4Q5 A0A131Y4F6 A0A1E1X2P5 A0A016UDM4 A0A131Y8X0 A0A147BJQ0 A0A016UEC4 A0A3B3RE95 A0A131Y3E3 A0A131Y3W4 W4YZP0 A0A131Y3C2 V5H708 A0A147BLD9 A0A131XRE4 K7EWV4 A0A2I0UP23 A0A3B3RWV8 W4Y8J8 W4ZK03 A0A091HUB5 A0A131Y4Q4 A0A1E1XND6 A0A2I0UDR0 W4ZIH4 K7EYK2 A0A091I8M3 K7EYD9 K7EY85 K7EYG1
Pubmed
EMBL
AB126052
BAD86652.1
AY359886
AAQ57129.1
RSAL01002067
RVE40593.1
+ More
RSAL01000164 RVE45343.1 RSAL01003136 RVE40332.1 RSAL01000002 RVE54942.1 FM995623 CAX36785.1 RVE55041.1 RVE55094.1 RSAL01000422 RVE41792.1 ODYU01007424 SOQ50185.1 ODYU01002162 SOQ39302.1 AMCI01002135 EJX03495.1 HAAD01001474 CDG67706.1 RSAL01000193 RVE44603.1 RSAL01000058 RVE49758.1 ODYU01000366 SOQ35027.1 DPOC01000269 HCX22453.1 RSAL01000030 RVE51708.1 LWDP01000391 ORD92879.1 GDRN01105059 JAI57791.1 RSAL01000110 RVE47159.1 ODYU01004409 SOQ44252.1 ODYU01011959 SOQ58001.1 ODYU01007423 SOQ50184.1 ODYU01006795 SOQ48973.1 ODYU01003793 SOQ42997.1 MRZV01001153 PIK40187.1 AAGJ04079835 GFAC01000919 JAT98269.1 GEFH01003922 JAP64659.1 GFAA01002385 JAU01050.1 MRZV01000414 PIK50568.1 AAGJ04112043 GBZX01002173 JAG90567.1 GEFM01001453 JAP74343.1 GEGO01004630 JAR90774.1 GFAA01000199 JAU03236.1 GFAA01002657 JAU00778.1 GFAA01002479 JAU00956.1 GADI01003787 JAA70021.1 KZ505886 PKU43598.1 GFAC01000877 JAT98311.1 NSIT01000239 PJE78188.1 GEFM01002424 JAP73372.1 GFAC01005665 JAT93523.1 JARK01001381 EYC12952.1 GEFM01000818 JAP74978.1 GEGO01004433 JAR90971.1 EYC12953.1 GEFM01002419 JAP73377.1 GEFM01001613 JAP74183.1 AAGJ04095885 GEFM01002439 JAP73357.1 GANP01005613 JAB78855.1 GEGO01003794 JAR91610.1 GEFM01005882 JAP69914.1 AGCU01000442 KZ505669 PKU47808.1 AAGJ04028723 AAGJ04028724 AAGJ04028725 AAGJ04028970 KL217824 KFO99526.1 GEFM01002324 JAP73472.1 GFAA01002660 JAU00775.1 KZ505846 PKU44169.1 AGCU01035999 KL218275 KFP03813.1 AGCU01094663 AGCU01082246 AGCU01122093
RSAL01000164 RVE45343.1 RSAL01003136 RVE40332.1 RSAL01000002 RVE54942.1 FM995623 CAX36785.1 RVE55041.1 RVE55094.1 RSAL01000422 RVE41792.1 ODYU01007424 SOQ50185.1 ODYU01002162 SOQ39302.1 AMCI01002135 EJX03495.1 HAAD01001474 CDG67706.1 RSAL01000193 RVE44603.1 RSAL01000058 RVE49758.1 ODYU01000366 SOQ35027.1 DPOC01000269 HCX22453.1 RSAL01000030 RVE51708.1 LWDP01000391 ORD92879.1 GDRN01105059 JAI57791.1 RSAL01000110 RVE47159.1 ODYU01004409 SOQ44252.1 ODYU01011959 SOQ58001.1 ODYU01007423 SOQ50184.1 ODYU01006795 SOQ48973.1 ODYU01003793 SOQ42997.1 MRZV01001153 PIK40187.1 AAGJ04079835 GFAC01000919 JAT98269.1 GEFH01003922 JAP64659.1 GFAA01002385 JAU01050.1 MRZV01000414 PIK50568.1 AAGJ04112043 GBZX01002173 JAG90567.1 GEFM01001453 JAP74343.1 GEGO01004630 JAR90774.1 GFAA01000199 JAU03236.1 GFAA01002657 JAU00778.1 GFAA01002479 JAU00956.1 GADI01003787 JAA70021.1 KZ505886 PKU43598.1 GFAC01000877 JAT98311.1 NSIT01000239 PJE78188.1 GEFM01002424 JAP73372.1 GFAC01005665 JAT93523.1 JARK01001381 EYC12952.1 GEFM01000818 JAP74978.1 GEGO01004433 JAR90971.1 EYC12953.1 GEFM01002419 JAP73377.1 GEFM01001613 JAP74183.1 AAGJ04095885 GEFM01002439 JAP73357.1 GANP01005613 JAB78855.1 GEGO01003794 JAR91610.1 GEFM01005882 JAP69914.1 AGCU01000442 KZ505669 PKU47808.1 AAGJ04028723 AAGJ04028724 AAGJ04028725 AAGJ04028970 KL217824 KFO99526.1 GEFM01002324 JAP73472.1 GFAA01002660 JAU00775.1 KZ505846 PKU44169.1 AGCU01035999 KL218275 KFP03813.1 AGCU01094663 AGCU01082246 AGCU01122093
Proteomes
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR016190 Transl_init_fac_IF2/IF5_Zn-bd
IPR002735 Transl_init_fac_IF2/IF5
IPR016189 Transl_init_fac_IF2/IF5_N
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR034653 SPF45_RRM
IPR003954 RRM_dom_euk
IPR000467 G_patch_dom
IPR040052 RBM17
IPR035979 RBD_domain_sf
IPR036322 WD40_repeat_dom_sf
IPR040382 NOL10/Enp2
IPR012580 NUC153
IPR001680 WD40_repeat
IPR000738 WHEP-TRS_dom
IPR009068 S15_NS1_RNA-bd
IPR036857 Thyroglobulin_1_sf
IPR000716 Thyroglobulin_1
IPR013083 Znf_RING/FYVE/PHD
IPR036987 SRA-YDG_sf
IPR018957 Znf_C3HC4_RING-type
IPR015947 PUA-like_sf
IPR017907 Znf_RING_CS
IPR001841 Znf_RING
IPR003105 SRA_YDG
IPR000719 Prot_kinase_dom
IPR008266 Tyr_kinase_AS
IPR020635 Tyr_kinase_cat_dom
IPR017441 Protein_kinase_ATP_BS
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR005828 MFS_sugar_transport-like
IPR031986 GD_N
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR016190 Transl_init_fac_IF2/IF5_Zn-bd
IPR002735 Transl_init_fac_IF2/IF5
IPR016189 Transl_init_fac_IF2/IF5_N
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR034653 SPF45_RRM
IPR003954 RRM_dom_euk
IPR000467 G_patch_dom
IPR040052 RBM17
IPR035979 RBD_domain_sf
IPR036322 WD40_repeat_dom_sf
IPR040382 NOL10/Enp2
IPR012580 NUC153
IPR001680 WD40_repeat
IPR000738 WHEP-TRS_dom
IPR009068 S15_NS1_RNA-bd
IPR036857 Thyroglobulin_1_sf
IPR000716 Thyroglobulin_1
IPR013083 Znf_RING/FYVE/PHD
IPR036987 SRA-YDG_sf
IPR018957 Znf_C3HC4_RING-type
IPR015947 PUA-like_sf
IPR017907 Znf_RING_CS
IPR001841 Znf_RING
IPR003105 SRA_YDG
IPR000719 Prot_kinase_dom
IPR008266 Tyr_kinase_AS
IPR020635 Tyr_kinase_cat_dom
IPR017441 Protein_kinase_ATP_BS
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR005828 MFS_sugar_transport-like
IPR031986 GD_N
SUPFAM
ProteinModelPortal
Q5KTM5
Q6UV17
A0A3S2LP03
A0A3S2P9Q3
A0A3S2NHP4
A0A3S2NMS5
+ More
B9WPT5 A0A3S2NMX8 A0A3S2LQP1 A0A2H1WAR9 A0A2H1VFZ3 J9GT67 T2M6M7 A0A3S2TFH4 A0A3S2PF93 A0A2H1V2F4 A0A3D5S1K1 A0A3S2PHM8 A0A1Y1S3P2 A0A0P4VXU0 A0A3S2LI48 A0A2H1VUV7 A0A2H1WY65 A0A2H1WCG9 A0A2H1W7F5 A0A2H1VQC3 A0A2G8JWN9 W4Z5Z2 A0A1E1XGQ7 A0A131XCJ7 A0A1E1XPS6 A0A2G8KRG4 W4XMI9 A0A0C9SB62 A0A131Y4D1 A0A147BJ69 A0A1E1XVB3 A0A1E1XN74 A0A1E1XNW6 A0A0K8RG16 A0A2I0UC10 A0A1E1XGZ3 A0A2H9T4Q5 A0A131Y4F6 A0A1E1X2P5 A0A016UDM4 A0A131Y8X0 A0A147BJQ0 A0A016UEC4 A0A3B3RE95 A0A131Y3E3 A0A131Y3W4 W4YZP0 A0A131Y3C2 V5H708 A0A147BLD9 A0A131XRE4 K7EWV4 A0A2I0UP23 A0A3B3RWV8 W4Y8J8 W4ZK03 A0A091HUB5 A0A131Y4Q4 A0A1E1XND6 A0A2I0UDR0 W4ZIH4 K7EYK2 A0A091I8M3 K7EYD9 K7EY85 K7EYG1
B9WPT5 A0A3S2NMX8 A0A3S2LQP1 A0A2H1WAR9 A0A2H1VFZ3 J9GT67 T2M6M7 A0A3S2TFH4 A0A3S2PF93 A0A2H1V2F4 A0A3D5S1K1 A0A3S2PHM8 A0A1Y1S3P2 A0A0P4VXU0 A0A3S2LI48 A0A2H1VUV7 A0A2H1WY65 A0A2H1WCG9 A0A2H1W7F5 A0A2H1VQC3 A0A2G8JWN9 W4Z5Z2 A0A1E1XGQ7 A0A131XCJ7 A0A1E1XPS6 A0A2G8KRG4 W4XMI9 A0A0C9SB62 A0A131Y4D1 A0A147BJ69 A0A1E1XVB3 A0A1E1XN74 A0A1E1XNW6 A0A0K8RG16 A0A2I0UC10 A0A1E1XGZ3 A0A2H9T4Q5 A0A131Y4F6 A0A1E1X2P5 A0A016UDM4 A0A131Y8X0 A0A147BJQ0 A0A016UEC4 A0A3B3RE95 A0A131Y3E3 A0A131Y3W4 W4YZP0 A0A131Y3C2 V5H708 A0A147BLD9 A0A131XRE4 K7EWV4 A0A2I0UP23 A0A3B3RWV8 W4Y8J8 W4ZK03 A0A091HUB5 A0A131Y4Q4 A0A1E1XND6 A0A2I0UDR0 W4ZIH4 K7EYK2 A0A091I8M3 K7EYD9 K7EY85 K7EYG1
Ontologies
Topology
Subcellular location
Nucleus
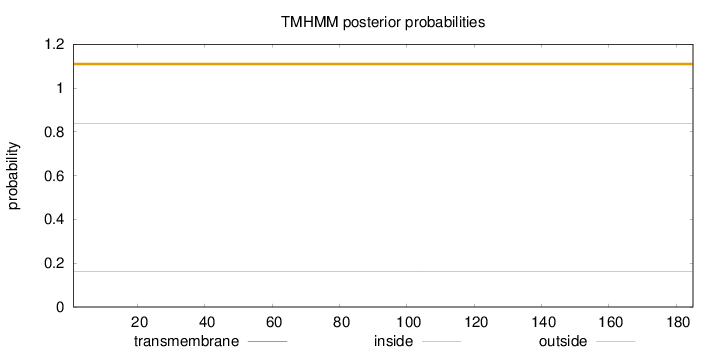
Length:
185
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000810000000000001
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.16227
outside
1 - 185
Population Genetic Test Statistics
Pi
12.550415
Theta
21.720932
Tajima's D
-1.111336
CLR
3.194049
CSRT
0.113244337783111
Interpretation
Uncertain
