Pre Gene Modal
BGIBMGA002823
Annotation
PREDICTED:_rab5_GDP/GTP_exchange_factor_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.726
Sequence
CDS
ATGCCTTCACTACGGATCGACCAAAACGACTTGAAGTGCAAGAATGGATGTGATTATTTTGGCAACCCACAATGGCAGGGGTACTGCTCGAAATGTCACAGGGAACAGATGCAGAGGCAAAGAAAAGCAGAGAAAGCCTCATCGTTTACATCGACTCTTCCTAGATCGGACCAGAGGAAGTCCGAACGCGGCCTCAAGTTGTCCTCGCACGCGTCCTTCTCGAAGTTCGAAGAGAAGCGCCTGCGACAAAGCGAAACGCTCAAGAAGACTAATCTACTGAAGTTTAACGTGTTCAAGAAGAATATTACAGATGACCATGAAATCTCAACGGAAAGACGTCAGCCAGAGTTTAAGATTCCGACCGCAGTGTTCGAAGGTATGAAGGCAGACTTCCGGGTCCGCTTTCCCTCGTTTCCCCCCCAGATAGACCGGGACGCTAGGGGCTTCGTTCACACCTTCATCCTGGATGTCATAAAATGGGCTGCTATAATGAATGTGGACGAGCTCTCGGAGAGGGTGCAGAAGCAGTACCAGCGCTTCATGAAGCACATGGAAACGTCGCAGCACTACGCATCTGTAGACTCGGACGCCAGAGAGCTGCTCATAGATTTCGTCGAGAAACACGCCATGACCTACTTACATGAGTTGCCGTCTATAGTGTTTTCGCCGAGTGGAACCGATGACGAACGCCAGGACCGCGCCATGTCGGAGCGGATACAGCAACTGAGCTGGGTGGGGGAAAAGCACCTCGAATGTAAACTGGACAGGAACAATGCTGAATGCCGACGGTTGCTCTACAAGGCTATAAGCGAGCTGCTGGGTATGGACGCGGCGCGCTGGTCGGGCGCCAAGCTGGCCCGCGTGCGGCGCTGCGCCCGCCACGTGCTGCGGCTGGCCGCGCCCGCGCCCGCCTCCGCCGACGACCTGCTGCCCGCGCTCATACTCGCCGTCCTGAAAGCCAACCCGCCACGTTTGATAAGCAACATTAATTACGTGACCCGCTTCTGCAACGCCCAGAGGCTGATGACCGGCGAGGGTGGCTACTACTTTACTAACTTGTGCTGTGCGGTGTCGTTCATCCAGAATATGACGGCGGAGTCTCTGAGCATGGACAAGAAGGAGTTCGACTGCTACATGGCGATGCCGGCCTCCATAGGCGGGAGTGCCTGGGCGGCCGCGCTGTCGCTGTGCGGCGCCACACGCGAGGCCGAGGAACAGAGCGCTCTAGCAGCGAAACTTCTAGAACAGACCAAAGAGATACAGAGGGAGGCCGAGAACATGGCGAACAACGCGGCTACATTTGAGGAACAAATCGCGAAGAAAGTTCAAGACGTTTTAGCAAAGACCCCACTGGAGATCAAACCACGGCGAGAACTGCCGCGTCTCAGACGCGAGGGCGTCGGGTCCGCGCCGCCGCCCACCACCGAGCCCGGCCCGGCGCGGACCTCAGAACCGCCAGCGCTGCTCAACGAGCCGAGCGTAGTCATCGACGAAACCGAAAAACTAAACATCTTAGGCTTCGAAGTGGTCGAGGAGCGGTCGCCGTCCAAGTCGCCGCAGCTGGACCGCCTCTCCGACCTCAAACAGAGCAGTTCAACGGAACTACTGACCCCATCCCCGATGGTCCTCACTCCGTTCGACTGCAGGTCCCTAGACGAGGTGCTGACCCCGGACGAGTTCCCGGACAACATCCCCGGACTGAGCGACGTCAATTATGACATAGACCTGTCAGATTTCAGCGCGGATACCAGTCTGGCGGAGGACCCCCCCGCCCCCGACCGCCGAAGGACCTTCGATCCATTCGCGCCTCAGACCTCAAAAGACTCGTACAAGTTGCTGGAGTCCTTTGATGAATTTAGCATCGTGGATAACGCGCGCGAGTCGGACCCCTACGAGGTCGTGCACAGGCCGGACCCGTTTGGTTCTGTTTTGGACAACGATGAGGGTAGAGCGTCCATTCTGGACGACAGCGGTTCCCCCACGGCCGCCTGCCTGCTGCCCTCACCGATACAGCCTCAAAACAGCGCGAAGCCAACTTAG
Protein
MPSLRIDQNDLKCKNGCDYFGNPQWQGYCSKCHREQMQRQRKAEKASSFTSTLPRSDQRKSERGLKLSSHASFSKFEEKRLRQSETLKKTNLLKFNVFKKNITDDHEISTERRQPEFKIPTAVFEGMKADFRVRFPSFPPQIDRDARGFVHTFILDVIKWAAIMNVDELSERVQKQYQRFMKHMETSQHYASVDSDARELLIDFVEKHAMTYLHELPSIVFSPSGTDDERQDRAMSERIQQLSWVGEKHLECKLDRNNAECRRLLYKAISELLGMDAARWSGAKLARVRRCARHVLRLAAPAPASADDLLPALILAVLKANPPRLISNINYVTRFCNAQRLMTGEGGYYFTNLCCAVSFIQNMTAESLSMDKKEFDCYMAMPASIGGSAWAAALSLCGATREAEEQSALAAKLLEQTKEIQREAENMANNAATFEEQIAKKVQDVLAKTPLEIKPRRELPRLRREGVGSAPPPTTEPGPARTSEPPALLNEPSVVIDETEKLNILGFEVVEERSPSKSPQLDRLSDLKQSSSTELLTPSPMVLTPFDCRSLDEVLTPDEFPDNIPGLSDVNYDIDLSDFSADTSLAEDPPAPDRRRTFDPFAPQTSKDSYKLLESFDEFSIVDNARESDPYEVVHRPDPFGSVLDNDEGRASILDDSGSPTAACLLPSPIQPQNSAKPT
Summary
Uniprot
H9IZY8
A0A2A4JI99
A0A194Q4A5
A0A0N1I6I1
A0A212ENU2
A0A0L7KCU0
+ More
A0A2J7PF72 A0A067RCD9 A0A1B6CM49 A0A1B6D297 V5GX42 A0A1Y1N3Y8 A0A0T6AU62 B0WNL8 A0A1Q3F7B1 N6UF97 A0A1Q3F7W5 A0A1Q3F5L0 A0A1Q3F757 U5EY36 B4L0N2 A0A1B6HC12 A0A0K8VH28 A0A0M5J0K7 A0A034W2E6 B4LG79 B4J2E7 A0A0A1XFM6 B4MXZ5 A0A1A9W4K5 A0A2J7PF74 Q9W0H9 A0A336K2I2 W8BK82 A0A336MKV9
A0A2J7PF72 A0A067RCD9 A0A1B6CM49 A0A1B6D297 V5GX42 A0A1Y1N3Y8 A0A0T6AU62 B0WNL8 A0A1Q3F7B1 N6UF97 A0A1Q3F7W5 A0A1Q3F5L0 A0A1Q3F757 U5EY36 B4L0N2 A0A1B6HC12 A0A0K8VH28 A0A0M5J0K7 A0A034W2E6 B4LG79 B4J2E7 A0A0A1XFM6 B4MXZ5 A0A1A9W4K5 A0A2J7PF74 Q9W0H9 A0A336K2I2 W8BK82 A0A336MKV9
Pubmed
EMBL
BABH01018930
BABH01018931
NWSH01001468
PCG71153.1
KQ459472
KPJ00179.1
+ More
KQ461186 KPJ07388.1 AGBW02013618 OWR43127.1 JTDY01010155 KOB60674.1 NEVH01026085 PNF14986.1 KK852556 KDR21427.1 GEDC01022772 JAS14526.1 GEDC01017613 JAS19685.1 GALX01002129 JAB66337.1 GEZM01013431 GEZM01013426 GEZM01013425 JAV92559.1 LJIG01022843 KRT78427.1 DS232013 EDS31854.1 GFDL01011652 JAV23393.1 APGK01024356 KB740544 KB632130 ENN80415.1 ERL88966.1 GFDL01011442 JAV23603.1 GFDL01012287 JAV22758.1 GFDL01011686 JAV23359.1 GANO01000556 JAB59315.1 CH933809 EDW18109.1 GECU01035498 JAS72208.1 GDHF01014474 JAI37840.1 CP012525 ALC42948.1 GAKP01010088 JAC48864.1 CH940647 EDW69387.1 CH916366 EDV96006.1 GBXI01004078 JAD10214.1 CH963876 EDW76984.2 PNF14981.1 AE014296 AY058763 AAF47467.1 AAL13992.1 UFQS01000091 UFQT01000091 SSW99184.1 SSX19566.1 GAMC01009122 JAB97433.1 UFQS01000704 UFQT01000704 SSX06314.1 SSX26668.1
KQ461186 KPJ07388.1 AGBW02013618 OWR43127.1 JTDY01010155 KOB60674.1 NEVH01026085 PNF14986.1 KK852556 KDR21427.1 GEDC01022772 JAS14526.1 GEDC01017613 JAS19685.1 GALX01002129 JAB66337.1 GEZM01013431 GEZM01013426 GEZM01013425 JAV92559.1 LJIG01022843 KRT78427.1 DS232013 EDS31854.1 GFDL01011652 JAV23393.1 APGK01024356 KB740544 KB632130 ENN80415.1 ERL88966.1 GFDL01011442 JAV23603.1 GFDL01012287 JAV22758.1 GFDL01011686 JAV23359.1 GANO01000556 JAB59315.1 CH933809 EDW18109.1 GECU01035498 JAS72208.1 GDHF01014474 JAI37840.1 CP012525 ALC42948.1 GAKP01010088 JAC48864.1 CH940647 EDW69387.1 CH916366 EDV96006.1 GBXI01004078 JAD10214.1 CH963876 EDW76984.2 PNF14981.1 AE014296 AY058763 AAF47467.1 AAL13992.1 UFQS01000091 UFQT01000091 SSW99184.1 SSX19566.1 GAMC01009122 JAB97433.1 UFQS01000704 UFQT01000704 SSX06314.1 SSX26668.1
Proteomes
SUPFAM
SSF109993
SSF109993
Gene 3D
ProteinModelPortal
H9IZY8
A0A2A4JI99
A0A194Q4A5
A0A0N1I6I1
A0A212ENU2
A0A0L7KCU0
+ More
A0A2J7PF72 A0A067RCD9 A0A1B6CM49 A0A1B6D297 V5GX42 A0A1Y1N3Y8 A0A0T6AU62 B0WNL8 A0A1Q3F7B1 N6UF97 A0A1Q3F7W5 A0A1Q3F5L0 A0A1Q3F757 U5EY36 B4L0N2 A0A1B6HC12 A0A0K8VH28 A0A0M5J0K7 A0A034W2E6 B4LG79 B4J2E7 A0A0A1XFM6 B4MXZ5 A0A1A9W4K5 A0A2J7PF74 Q9W0H9 A0A336K2I2 W8BK82 A0A336MKV9
A0A2J7PF72 A0A067RCD9 A0A1B6CM49 A0A1B6D297 V5GX42 A0A1Y1N3Y8 A0A0T6AU62 B0WNL8 A0A1Q3F7B1 N6UF97 A0A1Q3F7W5 A0A1Q3F5L0 A0A1Q3F757 U5EY36 B4L0N2 A0A1B6HC12 A0A0K8VH28 A0A0M5J0K7 A0A034W2E6 B4LG79 B4J2E7 A0A0A1XFM6 B4MXZ5 A0A1A9W4K5 A0A2J7PF74 Q9W0H9 A0A336K2I2 W8BK82 A0A336MKV9
PDB
4Q9U
E-value=1.84511e-37,
Score=393
Ontologies
GO
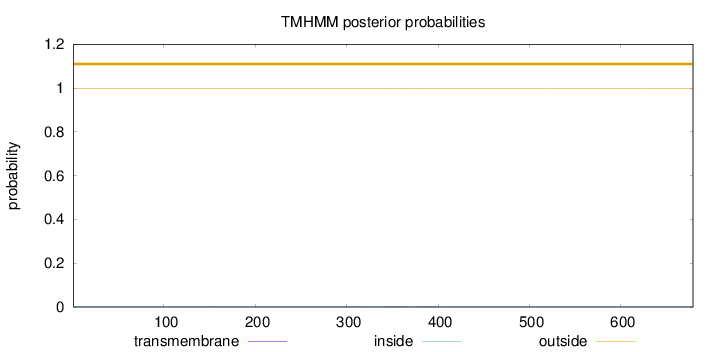
Topology
Length:
679
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08877
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00241
outside
1 - 679
Population Genetic Test Statistics
Pi
29.008098
Theta
27.433235
Tajima's D
0.054321
CLR
12.763411
CSRT
0.386480675966202
Interpretation
Uncertain
