Gene
KWMTBOMO06010
Annotation
PREDICTED:_alpha-ketoglutarate-dependent_dioxygenase_alkB_homolog_2-like_[Papilio_polytes]
Full name
DNA oxidative demethylase ALKBH2
Alternative Name
Alkylated DNA repair protein alkB homolog 2
Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2
Oxy DC1
Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2
Oxy DC1
Location in the cell
Cytoplasmic Reliability : 1.068 Mitochondrial Reliability : 1.593
Sequence
CDS
ATGGTATACAAATTGGTTTTTGGCAAAACGTATCCTTTGCCCAGACAACAAGTGGCATACGGAGATGATGGCATAACATACACATACTCAGGGGTTACAGTACCTGCCCTACCGTGGCCTGCACCTGTTCTCGCGTTAAGGGATTTTCTTTACGGAATAACGAATATCAGATATGATTTCGTGTTGATAAATAAATACAGAAACGGTATAGATCATATGGGCGAACATAGGGACAATGAACCAGAATTGGACCCAAGCTACCCCATCGCATCTATTTCTTTAGGACAGGAACGACCGTTTGTATTGAAACATAGAGATGCAAGAAAATCTGGAAAAGAGAAAAGGGCAATACCTACAGGTTTGTAG
Protein
MVYKLVFGKTYPLPRQQVAYGDDGITYTYSGVTVPALPWPAPVLALRDFLYGITNIRYDFVLINKYRNGIDHMGEHRDNEPELDPSYPIASISLGQERPFVLKHRDARKSGKEKRAIPTGL
Summary
Description
Dioxygenase that repairs alkylated DNA and RNA containing 1-methyladenine and 3-methylcytosine by oxidative demethylation. Can also repair alkylated DNA containing 1-ethenoadenine (in vitro). Has strong preference for double-stranded DNA. Has low efficiency with single-stranded substrates. Requires molecular oxygen, alpha-ketoglutarate and iron.
Catalytic Activity
2-oxoglutarate + a methylated nucleobase within DNA + O2 = a nucleobase within DNA + CO2 + formaldehyde + succinate
Cofactor
Fe(2+)
Similarity
Belongs to the alkB family.
Keywords
3D-structure
Alternative splicing
Complete proteome
Dioxygenase
DNA damage
DNA repair
Iron
Magnesium
Metal-binding
Nucleus
Oxidoreductase
Polymorphism
Reference proteome
Feature
chain DNA oxidative demethylase ALKBH2
splice variant In isoform 2.
sequence variant In dbSNP:rs33962311.
splice variant In isoform 2.
sequence variant In dbSNP:rs33962311.
Uniprot
A0A2A4JIB4
A0A2H1WQ15
A0A0N1PGK9
A0A194Q3R6
A0A1Y1MM59
A0A0T6B3S1
+ More
A0A1B6CYH9 A0A067RC09 D6X1E3 A0A2J7PF86 A0A2J7PF91 A0A0S7MGW3 A0A1A7YV10 A0A2I4BPZ0 M4AUZ1 A0A3P9H4P9 A0A3B5QHA3 A0A3B5Q4E6 A0A1A8NXQ1 H3DEL7 A0A3P9MCG8 A0A1A8PX10 A0A3Q3BCE2 A0A1A8AY50 A0A1A8IXR2 A0A3B3V2J7 A0A1A8G2W4 A0A3P9MCJ2 A0A2D0PP36 A0A1A8UZE3 A0A1A8C4P2 G3P2H6 A0A2D0PRQ7 A0A3B4YX32 A0A2H8TFP4 A0A2H8TL25 A0A3B4UDY2 A0A3B5KLA0 A0A3B3HKF1 A0A3B5K4V9 A0A3Q3MES8 G3P2H0 A0A3Q3JUL3 A0A3Q2PLH1 A0A2S2PBH9 H2LS59 A0A087Y0C3 I3K701 I3K702 A0A3Q1G2I5 A0A147A9H4 A0A2U9BSQ7 A0A3Q2FHA8 A0A3P9CEN5 A0A3Q1HWQ0 A0A3B3B8B5 A0A1B6ISZ2 A0A3P9QGC7 A0A3P8R9T4 A0A3B4GG59 A0A3N0Z0K3 A0A3B3XTA9 A0A3B3XSY1 A0A2U9BS92 A0A3Q0SIH0 C4WWX2 A0A2R5LHV2 A0A3Q3XIP8 A0A3P8U6V0 A0A060WH90 A0A3B1KAJ0 E9QG26 A0A3Q4H1K2 A0A3Q2UZE1 A3QK41 A0A2P2I833 A0A3Q1BN59 A0A3P8Y769 A0A0P4WCI8 G3R2E5 A0A2R9AWD5 H2Q6U0 Q6NS38 G3RZ41 A0A3B4CST3 A0A2G8JT43 A0A2K5HSH0 Q4RSX1 A0A2K6BD08 A0A2K6MNL8 A0A2K6PVN6 A0A2K5KWI2 A0A2K5XJD9 A0A096MYW1 G7PI61 F7DWK4 A0A2K5DI25 A0A2J8XLJ4 A0A3P8VI60 A0A3Q2YVB0
A0A1B6CYH9 A0A067RC09 D6X1E3 A0A2J7PF86 A0A2J7PF91 A0A0S7MGW3 A0A1A7YV10 A0A2I4BPZ0 M4AUZ1 A0A3P9H4P9 A0A3B5QHA3 A0A3B5Q4E6 A0A1A8NXQ1 H3DEL7 A0A3P9MCG8 A0A1A8PX10 A0A3Q3BCE2 A0A1A8AY50 A0A1A8IXR2 A0A3B3V2J7 A0A1A8G2W4 A0A3P9MCJ2 A0A2D0PP36 A0A1A8UZE3 A0A1A8C4P2 G3P2H6 A0A2D0PRQ7 A0A3B4YX32 A0A2H8TFP4 A0A2H8TL25 A0A3B4UDY2 A0A3B5KLA0 A0A3B3HKF1 A0A3B5K4V9 A0A3Q3MES8 G3P2H0 A0A3Q3JUL3 A0A3Q2PLH1 A0A2S2PBH9 H2LS59 A0A087Y0C3 I3K701 I3K702 A0A3Q1G2I5 A0A147A9H4 A0A2U9BSQ7 A0A3Q2FHA8 A0A3P9CEN5 A0A3Q1HWQ0 A0A3B3B8B5 A0A1B6ISZ2 A0A3P9QGC7 A0A3P8R9T4 A0A3B4GG59 A0A3N0Z0K3 A0A3B3XTA9 A0A3B3XSY1 A0A2U9BS92 A0A3Q0SIH0 C4WWX2 A0A2R5LHV2 A0A3Q3XIP8 A0A3P8U6V0 A0A060WH90 A0A3B1KAJ0 E9QG26 A0A3Q4H1K2 A0A3Q2UZE1 A3QK41 A0A2P2I833 A0A3Q1BN59 A0A3P8Y769 A0A0P4WCI8 G3R2E5 A0A2R9AWD5 H2Q6U0 Q6NS38 G3RZ41 A0A3B4CST3 A0A2G8JT43 A0A2K5HSH0 Q4RSX1 A0A2K6BD08 A0A2K6MNL8 A0A2K6PVN6 A0A2K5KWI2 A0A2K5XJD9 A0A096MYW1 G7PI61 F7DWK4 A0A2K5DI25 A0A2J8XLJ4 A0A3P8VI60 A0A3Q2YVB0
EC Number
1.14.11.33
Pubmed
26354079
28004739
24845553
18362917
19820115
23542700
+ More
17554307 15496914 21551351 25186727 29451363 24755649 25329095 23594743 25069045 22398555 22722832 16136131 17979886 16541075 15489334 12486230 12594517 16174769 18519673 21269460 18432238 20223766 29023486 25362486 22002653 17431167 25319552 24487278
17554307 15496914 21551351 25186727 29451363 24755649 25329095 23594743 25069045 22398555 22722832 16136131 17979886 16541075 15489334 12486230 12594517 16174769 18519673 21269460 18432238 20223766 29023486 25362486 22002653 17431167 25319552 24487278
EMBL
NWSH01001468
PCG71152.1
ODYU01010151
SOQ55066.1
KQ461186
KPJ07389.1
+ More
KQ459472 KPJ00177.1 GEZM01029000 JAV86168.1 LJIG01015997 KRT81924.1 GEDC01018769 GEDC01009461 JAS18529.1 JAS27837.1 KK852556 KDR21426.1 KQ971372 EFA10619.1 NEVH01026085 PNF14988.1 PNF14987.1 GBYX01028228 JAP00977.1 HADX01011548 SBP33780.1 HAEG01005309 SBR73806.1 HAEH01009002 SBR86010.1 HADY01020926 SBP59411.1 HAED01015682 SBR02127.1 HAEB01019030 SBQ65557.1 HAEJ01012509 SBS52966.1 HADZ01010931 SBP74872.1 GFXV01001076 MBW12881.1 GFXV01002944 MBW14749.1 GGMR01013657 MBY26276.1 AYCK01007732 AERX01026848 GCES01011162 JAR75161.1 CP026251 AWP07081.1 GECU01017716 JAS89990.1 RJVU01018862 ROL51428.1 AWP07078.1 AWP07079.1 AWP07080.1 ABLF02039828 AK342327 BAH72392.1 GGLE01004984 MBY09110.1 FR904540 CDQ66366.1 CR848794 CR931763 IACF01004580 LAB70169.1 GDRN01063675 GDRN01063673 JAI64942.1 CABD030086578 CABD030086579 AJFE02116885 AC193909 GABC01002660 GABF01008002 NBAG03000240 JAA08678.1 JAA14143.1 PNI64854.1 PNI64857.1 AY754389 AB277859 AC011596 BC070489 MRZV01001305 PIK38879.1 CAAE01014999 CAG08511.1 AHZZ02002947 AQIA01012618 CM001286 EHH66688.1 JSUE03007717 JU322687 JV044372 CM001263 AFE66443.1 AFI34443.1 EHH21155.1 NDHI03003364 PNJ82904.1 PNJ82907.1
KQ459472 KPJ00177.1 GEZM01029000 JAV86168.1 LJIG01015997 KRT81924.1 GEDC01018769 GEDC01009461 JAS18529.1 JAS27837.1 KK852556 KDR21426.1 KQ971372 EFA10619.1 NEVH01026085 PNF14988.1 PNF14987.1 GBYX01028228 JAP00977.1 HADX01011548 SBP33780.1 HAEG01005309 SBR73806.1 HAEH01009002 SBR86010.1 HADY01020926 SBP59411.1 HAED01015682 SBR02127.1 HAEB01019030 SBQ65557.1 HAEJ01012509 SBS52966.1 HADZ01010931 SBP74872.1 GFXV01001076 MBW12881.1 GFXV01002944 MBW14749.1 GGMR01013657 MBY26276.1 AYCK01007732 AERX01026848 GCES01011162 JAR75161.1 CP026251 AWP07081.1 GECU01017716 JAS89990.1 RJVU01018862 ROL51428.1 AWP07078.1 AWP07079.1 AWP07080.1 ABLF02039828 AK342327 BAH72392.1 GGLE01004984 MBY09110.1 FR904540 CDQ66366.1 CR848794 CR931763 IACF01004580 LAB70169.1 GDRN01063675 GDRN01063673 JAI64942.1 CABD030086578 CABD030086579 AJFE02116885 AC193909 GABC01002660 GABF01008002 NBAG03000240 JAA08678.1 JAA14143.1 PNI64854.1 PNI64857.1 AY754389 AB277859 AC011596 BC070489 MRZV01001305 PIK38879.1 CAAE01014999 CAG08511.1 AHZZ02002947 AQIA01012618 CM001286 EHH66688.1 JSUE03007717 JU322687 JV044372 CM001263 AFE66443.1 AFI34443.1 EHH21155.1 NDHI03003364 PNJ82904.1 PNJ82907.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000027135
UP000007266
UP000235965
+ More
UP000192220 UP000002852 UP000265200 UP000007303 UP000265180 UP000264800 UP000261500 UP000221080 UP000007635 UP000261360 UP000261420 UP000005226 UP000001038 UP000261640 UP000261600 UP000265000 UP000028760 UP000005207 UP000257200 UP000246464 UP000265020 UP000265160 UP000265040 UP000261560 UP000242638 UP000265100 UP000261460 UP000261480 UP000261340 UP000007819 UP000261620 UP000265080 UP000193380 UP000018467 UP000000437 UP000261580 UP000264840 UP000257160 UP000265140 UP000001519 UP000240080 UP000002277 UP000005640 UP000261440 UP000230750 UP000233080 UP000233120 UP000233180 UP000233200 UP000233060 UP000233140 UP000028761 UP000009130 UP000233100 UP000006718 UP000233020 UP000265120 UP000264820
UP000192220 UP000002852 UP000265200 UP000007303 UP000265180 UP000264800 UP000261500 UP000221080 UP000007635 UP000261360 UP000261420 UP000005226 UP000001038 UP000261640 UP000261600 UP000265000 UP000028760 UP000005207 UP000257200 UP000246464 UP000265020 UP000265160 UP000265040 UP000261560 UP000242638 UP000265100 UP000261460 UP000261480 UP000261340 UP000007819 UP000261620 UP000265080 UP000193380 UP000018467 UP000000437 UP000261580 UP000264840 UP000257160 UP000265140 UP000001519 UP000240080 UP000002277 UP000005640 UP000261440 UP000230750 UP000233080 UP000233120 UP000233180 UP000233200 UP000233060 UP000233140 UP000028761 UP000009130 UP000233100 UP000006718 UP000233020 UP000265120 UP000264820
PRIDE
Pfam
PF13532 2OG-FeII_Oxy_2
Gene 3D
ProteinModelPortal
A0A2A4JIB4
A0A2H1WQ15
A0A0N1PGK9
A0A194Q3R6
A0A1Y1MM59
A0A0T6B3S1
+ More
A0A1B6CYH9 A0A067RC09 D6X1E3 A0A2J7PF86 A0A2J7PF91 A0A0S7MGW3 A0A1A7YV10 A0A2I4BPZ0 M4AUZ1 A0A3P9H4P9 A0A3B5QHA3 A0A3B5Q4E6 A0A1A8NXQ1 H3DEL7 A0A3P9MCG8 A0A1A8PX10 A0A3Q3BCE2 A0A1A8AY50 A0A1A8IXR2 A0A3B3V2J7 A0A1A8G2W4 A0A3P9MCJ2 A0A2D0PP36 A0A1A8UZE3 A0A1A8C4P2 G3P2H6 A0A2D0PRQ7 A0A3B4YX32 A0A2H8TFP4 A0A2H8TL25 A0A3B4UDY2 A0A3B5KLA0 A0A3B3HKF1 A0A3B5K4V9 A0A3Q3MES8 G3P2H0 A0A3Q3JUL3 A0A3Q2PLH1 A0A2S2PBH9 H2LS59 A0A087Y0C3 I3K701 I3K702 A0A3Q1G2I5 A0A147A9H4 A0A2U9BSQ7 A0A3Q2FHA8 A0A3P9CEN5 A0A3Q1HWQ0 A0A3B3B8B5 A0A1B6ISZ2 A0A3P9QGC7 A0A3P8R9T4 A0A3B4GG59 A0A3N0Z0K3 A0A3B3XTA9 A0A3B3XSY1 A0A2U9BS92 A0A3Q0SIH0 C4WWX2 A0A2R5LHV2 A0A3Q3XIP8 A0A3P8U6V0 A0A060WH90 A0A3B1KAJ0 E9QG26 A0A3Q4H1K2 A0A3Q2UZE1 A3QK41 A0A2P2I833 A0A3Q1BN59 A0A3P8Y769 A0A0P4WCI8 G3R2E5 A0A2R9AWD5 H2Q6U0 Q6NS38 G3RZ41 A0A3B4CST3 A0A2G8JT43 A0A2K5HSH0 Q4RSX1 A0A2K6BD08 A0A2K6MNL8 A0A2K6PVN6 A0A2K5KWI2 A0A2K5XJD9 A0A096MYW1 G7PI61 F7DWK4 A0A2K5DI25 A0A2J8XLJ4 A0A3P8VI60 A0A3Q2YVB0
A0A1B6CYH9 A0A067RC09 D6X1E3 A0A2J7PF86 A0A2J7PF91 A0A0S7MGW3 A0A1A7YV10 A0A2I4BPZ0 M4AUZ1 A0A3P9H4P9 A0A3B5QHA3 A0A3B5Q4E6 A0A1A8NXQ1 H3DEL7 A0A3P9MCG8 A0A1A8PX10 A0A3Q3BCE2 A0A1A8AY50 A0A1A8IXR2 A0A3B3V2J7 A0A1A8G2W4 A0A3P9MCJ2 A0A2D0PP36 A0A1A8UZE3 A0A1A8C4P2 G3P2H6 A0A2D0PRQ7 A0A3B4YX32 A0A2H8TFP4 A0A2H8TL25 A0A3B4UDY2 A0A3B5KLA0 A0A3B3HKF1 A0A3B5K4V9 A0A3Q3MES8 G3P2H0 A0A3Q3JUL3 A0A3Q2PLH1 A0A2S2PBH9 H2LS59 A0A087Y0C3 I3K701 I3K702 A0A3Q1G2I5 A0A147A9H4 A0A2U9BSQ7 A0A3Q2FHA8 A0A3P9CEN5 A0A3Q1HWQ0 A0A3B3B8B5 A0A1B6ISZ2 A0A3P9QGC7 A0A3P8R9T4 A0A3B4GG59 A0A3N0Z0K3 A0A3B3XTA9 A0A3B3XSY1 A0A2U9BS92 A0A3Q0SIH0 C4WWX2 A0A2R5LHV2 A0A3Q3XIP8 A0A3P8U6V0 A0A060WH90 A0A3B1KAJ0 E9QG26 A0A3Q4H1K2 A0A3Q2UZE1 A3QK41 A0A2P2I833 A0A3Q1BN59 A0A3P8Y769 A0A0P4WCI8 G3R2E5 A0A2R9AWD5 H2Q6U0 Q6NS38 G3RZ41 A0A3B4CST3 A0A2G8JT43 A0A2K5HSH0 Q4RSX1 A0A2K6BD08 A0A2K6MNL8 A0A2K6PVN6 A0A2K5KWI2 A0A2K5XJD9 A0A096MYW1 G7PI61 F7DWK4 A0A2K5DI25 A0A2J8XLJ4 A0A3P8VI60 A0A3Q2YVB0
PDB
3RZM
E-value=3.99349e-24,
Score=269
Ontologies
GO
Topology
Subcellular location
Nucleus
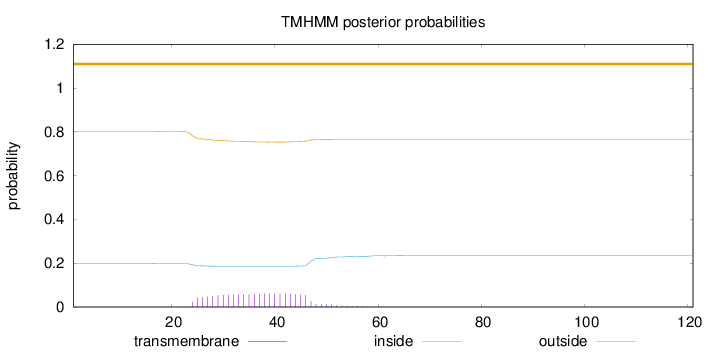
Length:
121
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.37783
Exp number, first 60 AAs:
1.37464
Total prob of N-in:
0.19904
outside
1 - 121
Population Genetic Test Statistics
Pi
18.04247
Theta
17.639114
Tajima's D
-0.197785
CLR
0.003427
CSRT
0.322033898305085
Interpretation
Uncertain
